 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do017188.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 416aa MW: 45883.1 Da PI: 9.0293 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 35.5 | 2.1e-11 | 132 | 161 | 5 | 34 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeek 34
k rr+++NReAAr+sR+RKka+i++Le+
Do017188.1 132 KTLRRLAQNREAARKSRLRKKAYIQQLETS 161
788************************974 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 8.9E-6 | 128 | 183 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 8.829 | 130 | 173 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.5E-8 | 130 | 172 | No hit | No description |
| Pfam | PF00170 | 1.8E-8 | 132 | 161 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.17E-7 | 132 | 166 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 135 | 150 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 4.9E-27 | 217 | 287 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 416 aa Download sequence Send to blast |
MTASSTMSKE SASYDMAEFD QSAIFLYLDG HDQEQRQTLN IFPSQPMHVA EPFPAKGVSM 60 GMVAAMLPNG NSSSPKRQEQ GGQRSAAPAP GPTVPLPNNS AKETRSSLTK KEATSGGKGA 120 TSGDQERVRD PKTLRRLAQN REAARKSRLR KKAYIQQLET SRIRLSQLEQ QVQVARVQGV 180 FLGTGEQPGF PSAPSPAAVF DMEYGRWVEE HSKLVFQLRA ALNEHLADEQ LQGFVNGAMA 240 QHEELLNLKG AMARADVFHL LSGLWASPTE RCFLWLGGFR PSEVIKVTLK HVEPLSEGQI 300 LGIYNLQQSV QEREEALNHS MEATQQSISD IVAAPDVAPA TFMGHMSLAM NKVASMEGFV 360 MRADGLRQQT LHKLHQILTT RQAARCMVAI ADYFHRLRAL STLWVARPRQ EDGPGL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator involved in defense response. {ECO:0000250|UniProtKB:Q7X993}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
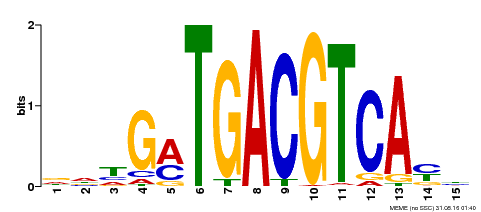
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do017188.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012698770.1 | 0.0 | transcription factor TGAL7 isoform X2 | ||||
| Swissprot | Q6H434 | 0.0 | TGAL7_ORYSJ; Transcription factor TGAL7 | ||||
| TrEMBL | A0A1E5WJ13 | 0.0 | A0A1E5WJ13_9POAL; TGACG-sequence-specific DNA-binding protein TGA-2.1 | ||||
| STRING | Si029966m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2732 | 36 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.3 | 1e-139 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



