 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do018432.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 850aa MW: 90260 Da PI: 6.2693 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 68 | 1.2e-21 | 111 | 166 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l+L+ rqVk+WFqNrR+++k
Do018432.1 111 KKRYHRHTPQQIQELEAVFKECPHPDEKQRMELSKRLNLESRQVKFWFQNRRTQMK 166
688999***********************************************999 PP
| |||||||
| 2 | START | 152.8 | 2.8e-48 | 329 | 564 | 4 | 205 |
HHHHHHHHHHHHHC-TT-EEEE.......EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEEC CS
START 4 eeaaqelvkkalaeepgWvkss.......esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevis 86
++a++el+k + +ep+W + +n de+ + f++ + + +ea r+ gv + +++lv +l+d + +W+e+++ +a+t++ is
Do018432.1 329 LAAMEELMKVSQMDEPLWLPTGpdggggfDTLNFDEYSRAFARVFGpspagYVSEATREAGVAITSSVDLVDSLMDAA-RWSEMFPcivaRASTTDIIS 426
789****************9989******************775559*******************************.******************** PP
TT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE............-TTS--....-TTSEE-EESSEEEEEEEECTCEEE CS
START 87 sg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvd............seqkppe...sssvvRaellpSgiliepksnghsk 163
sg g +qlm aelq+lsplvp R+++f+R+++q+ +g w++vdvSvd + q + ++++ ++llp+g++++++ ng+sk
Do018432.1 427 SGmggtrsGSIQLMHAELQVLSPLVPiREVLFLRFCKQHAEGLWAVVDVSVDailrpdggslhqHAQ---NggsAAGYMGCRLLPTGCIVQDMNNGYSK 522
***************************************************9444444444322233...245699*********************** PP
EEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 164 vtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqce 205
vtwv h+++++ +h l+r+l++sg+a ga++w+a lqrqc+
Do018432.1 523 VTWVVHAEYDETAVHHLYRPLLRSGQALGARRWLASLQRQCQ 564
*****************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.84E-22 | 93 | 168 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-23 | 98 | 168 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.301 | 108 | 168 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 8.7E-20 | 109 | 172 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.56E-20 | 111 | 168 | No hit | No description |
| Pfam | PF00046 | 4.0E-19 | 111 | 166 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 143 | 166 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 39.183 | 317 | 568 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.38E-25 | 321 | 564 | No hit | No description |
| CDD | cd08875 | 3.58E-108 | 323 | 564 | No hit | No description |
| SMART | SM00234 | 8.4E-34 | 326 | 565 | IPR002913 | START domain |
| Pfam | PF01852 | 2.4E-40 | 329 | 564 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.92E-14 | 640 | 840 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 850 aa Download sequence Send to blast |
MFDGAGSGVF SYDAAATAAA GAGMHNPARL IPAPPLPKPA GFGAPGLSLG LLGDMSRMGL 60 MGGGGSGSAG EGDSLGRGRE DENDSRSGSD NVDGASGDEL DPDNSNPRKK KKRYHRHTPQ 120 QIQELEAVFK ECPHPDEKQR MELSKRLNLE SRQVKFWFQN RRTQMKTQIE RHENALLRQE 180 NDKLRAENMT IREAMRNPIC ASCGGAAVLG EVSLEEQHLR IENARLKDEL DRVCALAGKF 240 LGRPISSGSP MSLQGCSGLE LGVGTNGAFG LGPLGASALQ PLPDLMGAGL PGPVGSAAMR 300 LPAGIGGLDG TMHGAADGID RTVLLELGLA AMEELMKVSQ MDEPLWLPTG PDGGGGFDTL 360 NFDEYSRAFA RVFGPSPAGY VSEATREAGV AITSSVDLVD SLMDAARWSE MFPCIVARAS 420 TTDIISSGMG GTRSGSIQLM HAELQVLSPL VPIREVLFLR FCKQHAEGLW AVVDVSVDAI 480 LRPDGGSLHQ HAQNGGSAAG YMGCRLLPTG CIVQDMNNGY SKVTWVVHAE YDETAVHHLY 540 RPLLRSGQAL GARRWLASLQ RQCQYLAILC SNSLPARDHA AITPVGRRSM LKLAQRMTDN 600 FCAGVCASAA QKWRRLDEWR GNGEGAGSAG NSAAGEGEEK VRMMARQSVG APGEPPGVVL 660 SATTSVRLSA TSPQRVFDYL RDEQRRGEWD ILANGEAMQE MDHIAKGQHH GNAVSLLRPN 720 ATSGNQNNML ILQETCTDSS GSLVVYAPVD VQSMHVVMNG GDSAYVSLLP SGFAILPDGH 780 SQPSNTAQGS PNGQSATNNT GSLVTVAFQI LVNNLPTAKL TVESVETVSN LLSCTIQKIK 840 SALQVSIVTP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 107 | 112 | RKKKKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
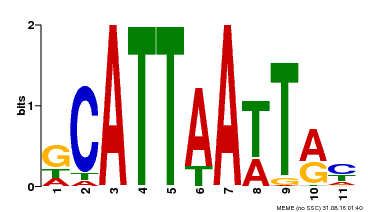
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do018432.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ250985 | 0.0 | AJ250985.1 Zea mays mRNA for OCL3 protein (ocl3 gene). | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025801167.1 | 0.0 | homeobox-leucine zipper protein ROC6-like | ||||
| Swissprot | Q7Y0V7 | 0.0 | ROC6_ORYSJ; Homeobox-leucine zipper protein ROC6 | ||||
| TrEMBL | A0A1E5V3Q9 | 0.0 | A0A1E5V3Q9_9POAL; Homeobox-leucine zipper protein ROC6 | ||||
| STRING | Sb02g030470.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1203 | 37 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||



