 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do021609.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 460aa MW: 51422.9 Da PI: 4.6937 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 111.1 | 7.9e-35 | 18 | 109 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellekik 98
Fl k+ye++ed+++++++sw g sfvv+++ +f++++LpkyFkh+nf+SF+RQLn+YgF+k++ e+ weF+++ F +g+++ll++i+
Do021609.1 18 FLIKTYEMVEDPATNHVVSWGPGGASFVVWNPPDFSRDLLPKYFKHNNFSSFIRQLNTYGFRKIDPER---------WEFANEDFIRGHTHLLKNIH 105
9****************************************************************999.........******************** PP
XXXX CS
HSF_DNA-bind 99 rkks 102
r+k
Do021609.1 106 RRKP 109
*985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46785 | 2.31E-36 | 13 | 107 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 9.9E-63 | 14 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene3D | G3DSA:1.10.10.10 | 6.9E-39 | 14 | 107 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 8.5E-20 | 18 | 41 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 6.9E-31 | 18 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 8.5E-20 | 56 | 68 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 57 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 8.5E-20 | 69 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 460 aa Download sequence Send to blast |
MEGSSGGAGG GPSSPPPFLI KTYEMVEDPA TNHVVSWGPG GASFVVWNPP DFSRDLLPKY 60 FKHNNFSSFI RQLNTYGFRK IDPERWEFAN EDFIRGHTHL LKNIHRRKPV HSHSLQTQVN 120 GPLAESERRE LEDEINRLKY EKSLLLADLQ RQNQQQYGIN WQMQSLEDRL VQMEQRQRNI 180 VASLCDILQR HGVVSGLILE RDHFSKKRRV PKIDFFNDEP TVEEQQVPYL KTLGAETPSM 240 SPIHLINAEP FEKIELALVS LENFFQRASH ANAEDMYSGP AEPSPALTLG EMNSAPMDTN 300 INQNSSAGLN PFSSTAGHAH LPCPLAESLS FVQSPMLTLA DLHEDADRTA EVDMNSETTT 360 GDTSQETTSE TGGSHMPAKV NDVFWERFLT GEAESGRQHA DDKSEATEAK EDMKIGIDCS 420 SLNHQNNVDQ ITEQMGYLDS TENGSEAAGN DLYHTICADP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 5e-26 | 1 | 107 | 12 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 5e-26 | 1 | 107 | 12 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 5e-26 | 1 | 107 | 12 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
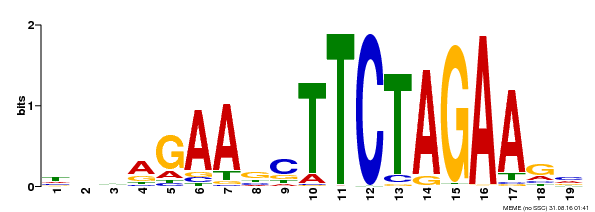
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do021609.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat stress. {ECO:0000269|PubMed:12032317}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004961415.1 | 0.0 | heat stress transcription factor A-4d | ||||
| Swissprot | Q93VB5 | 0.0 | HFA4D_ORYSJ; Heat stress transcription factor A-4d | ||||
| TrEMBL | A0A1E5VPP7 | 0.0 | A0A1E5VPP7_9POAL; Heat stress transcription factor A-4d | ||||
| STRING | Si022026m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3296 | 37 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 2e-71 | heat shock transcription factor A4A | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



