 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dusal.0015s00002.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Dunaliellaceae; Dunaliella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 504aa MW: 54670.4 Da PI: 5.7698 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.5 | 5.6e-17 | 112 | 157 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+WT eEde lv++v ++G ++W+ ++++ gR++k+c++rw++
Dusal.0015s00002.1.p 112 KGPWTHEEDEHLVRLVDKFGARNWSGMSKHIK-GRSGKSCRLRWLNQ 157
79*****************************9.***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 56.2 | 7.9e-18 | 164 | 208 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g++T+eEd ++ ++k +G++ W+tIa+ ++ gRt++ +k++w++
Dusal.0015s00002.1.p 164 KGPFTAEEDRSIIALHKIHGNK-WATIAKNLP-GRTDNAVKNHWNST 208
79********************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.468 | 107 | 158 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.6E-14 | 111 | 160 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.91E-30 | 111 | 205 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.7E-16 | 112 | 157 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-25 | 113 | 165 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.20E-12 | 114 | 156 | No hit | No description |
| PROSITE profile | PS51294 | 28.242 | 159 | 213 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.5E-17 | 163 | 211 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-15 | 164 | 208 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-25 | 166 | 212 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.53E-13 | 166 | 209 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 504 aa Download sequence Send to blast |
MDGFNIDTWL ASNAFAGQAV QPVAPQPAVD PQLFAQLFAN MGQQGVMGDD MGGALQQHTQ 60 HEQSRATPNY DSDEVELEDD EELGGSGGSG RGRGGRRGGG GGGGGGGEDS NKGPWTHEED 120 EHLVRLVDKF GARNWSGMSK HIKGRSGKSC RLRWLNQLNP NVKKGPFTAE EDRSIIALHK 180 IHGNKWATIA KNLPGRTDNA VKNHWNSTLR RRENEDVGPF VPLAGASLED ESRGGMDDVL 240 KSLMQQSSTG HANPNIMAAY DPNANAGYMQ PKDEEDTEDD GQGHKRRRYN GPDAIDPADS 300 PLRSLINSAV REVLAREMPL ILQMFSRTLK QSLAAQPRME LPGLLAALDT YINTAVAHRT 360 EQHLGTSQPG SQPQQQQQQQ QLALPAPLAA AAGAPPAGGV SSPQDLMQLL QQHLPPSEDA 420 LNPDFTSLLF GVALQQQQQQ QQQHLQQPQG LQQAVHGHEQ HQPQQQQHHD AVQAVAAMLG 480 GGQQQQQQLQ LLQQELKQEP QAQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-41 | 106 | 212 | 1 | 107 | B-MYB |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 93 | 101 | GGRRGGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00337 | DAP | Transfer from AT3G09230 | Download |
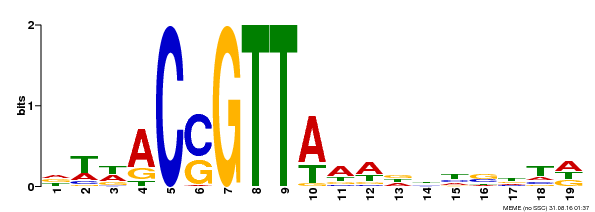
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09230.1 | 1e-44 | myb domain protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Dusal.0015s00002.1.p |



