 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dusal.0500s00002.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Dunaliellaceae; Dunaliella
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 272aa MW: 28124.9 Da PI: 10.2428 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.8 | 3.2e-19 | 125 | 173 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkr..krfslgkfgtaeeAakaaiaarkkleg 55
y+G+r+++ +g+W+AeIrdp + +r +lg+f+taeeAa+a++aa ++++g
Dusal.0500s00002.1.p 125 YRGIRQRP-WGKWAAEIRDP-----TagARRWLGTFDTAEEAARAYDAASRQIRG 173
9*******.**********9.....446**********************99998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 7.6E-31 | 124 | 181 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.7E-33 | 124 | 187 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 9.81E-22 | 124 | 182 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.341 | 124 | 181 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 3.4E-12 | 125 | 173 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-11 | 125 | 136 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.05E-28 | 125 | 181 | No hit | No description |
| PRINTS | PR00367 | 2.4E-11 | 147 | 163 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0034059 | Biological Process | response to anoxia | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MPTPPSPNDP HDPTDPNGFN EHEHRQQSQE QQQPVLPGGQ HPQSRRTASR NAAALLASAA 60 ADLDGDEASA SDSSETGKKR GASKGKARKP SANPPGNCAH PPPGGHLTGA VPPDQIKAKG 120 PNKTYRGIRQ RPWGKWAAEI RDPTAGARRW LGTFDTAEEA ARAYDAASRQ IRGPAAKCNF 180 PLPGETGAGR GTGARPDAKK VPAAANAAAA AAAQGTGLQG SHAPGPGVHG CAQKRARRTP 240 PPQQQQQQCF APGGPGLETS WSLLSDSFSQ D* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 5e-21 | 118 | 187 | 1 | 69 | ATERF1 |
| 3gcc_A | 5e-21 | 118 | 187 | 1 | 69 | ATERF1 |
| 5wx9_A | 7e-21 | 125 | 189 | 15 | 80 | Ethylene-responsive transcription factor ERF096 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
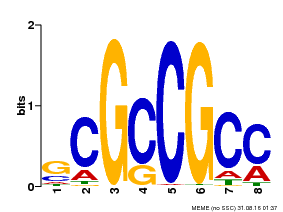
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP132 | 15 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50080.1 | 2e-19 | ethylene response factor 110 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Dusal.0500s00002.1.p |



