 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dusal.2891s00001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Dunaliellaceae; Dunaliella
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 264aa MW: 27469.9 Da PI: 10.0996 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.9 | 7.1e-20 | 190 | 238 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkr..krfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+W+AeIrdp + +r +lg+f+taeeAa+a++aa ++++g
Dusal.2891s00001.1.p 190 YRGVRQRP-WGKWAAEIRDP-----TagARRWLGTFDTAEEAARAYDAASRQIRG 238
9*******.**********9.....446**********************99998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.5E-22 | 189 | 247 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 3.2E-31 | 189 | 246 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.3E-34 | 189 | 252 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.525 | 189 | 246 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.45E-31 | 190 | 246 | No hit | No description |
| PRINTS | PR00367 | 1.7E-11 | 190 | 201 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.8E-13 | 190 | 238 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-11 | 212 | 228 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 264 aa Download sequence Send to blast |
MDSTGHQNPF QGWVHPVLPV PPLPPTKGAS PLPPPVSSNG YSGIPSVLPP RNTASQLAPP 60 PPAAAYAAYA ANMPYAQMPA YAPAGGAPSL ASLQAASTPA LDSPLQTAPP PPQQHSRRAA 120 AAAARATVVA AAADEEASST DSGDFAERKP VRKKKPVKKE TPIKQHDESI VSTGPPAPDV 180 IYSKGPNKTY RGVRQRPWGK WAAEIRDPTA GARRWLGTFD TAEEAARAYD AASRQIRGPA 240 ARCNFPLPGE VSAAQVSMVS VDK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-20 | 188 | 247 | 4 | 64 | ATERF1 |
| 3gcc_A | 1e-20 | 188 | 247 | 4 | 64 | ATERF1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00542 | DAP | Transfer from AT5G44210 | Download |
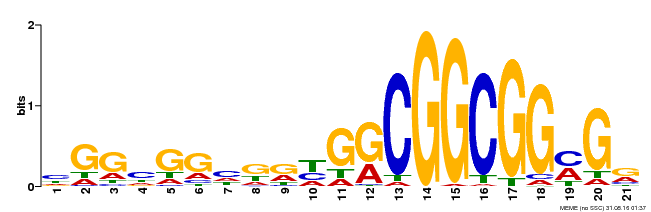
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP132 | 15 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G33710.1 | 4e-19 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Dusal.2891s00001.1.p |



