 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT09592 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 273aa MW: 31039.8 Da PI: 7.8038 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.6 | 1.2e-19 | 64 | 115 | 5 | 56 |
SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 5 ttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ eq+++Le+ Fe +++++ e+++++A+ lgL rqV vWFqNrRa++k
EMT09592 64 RRLALEQVRALERSFETDNKLDPERKARIARDLGLHPRQVAVWFQNRRARWK 115
467789*********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.4 | 2.9e-41 | 62 | 151 | 2 | 91 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
kkrrl+ eqv++LE+sFe+++kL+perK+++ar+Lgl+prqvavWFqnrRAR+ktkqlE+d++aL++++dal+++ ++L++++++L++e+
EMT09592 62 KKRRLALEQVRALERSFETDNKLDPERKARIARDLGLHPRQVAVWFQNRRARWKTKQLERDFNALRARHDALRADCDALRRDKDALAAEI 151
9*************************************************************************************9886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00389 | 1.9E-19 | 60 | 121 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.35E-18 | 60 | 119 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.552 | 62 | 117 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.74E-16 | 62 | 118 | No hit | No description |
| Pfam | PF00046 | 7.0E-17 | 64 | 115 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-19 | 69 | 116 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 3.5E-5 | 88 | 97 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 92 | 115 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 3.5E-5 | 97 | 113 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 6.2E-13 | 117 | 151 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MKRQIKRPTR SQESPEPGEK LAFAEKENLL AAYMEHDEPE LEEADEEEEE EERAMSCGLG 60 GKKRRLALEQ VRALERSFET DNKLDPERKA RIARDLGLHP RQVAVWFQNR RARWKTKQLE 120 RDFNALRARH DALRADCDAL RRDKDALAAE IFFRKLAFLR PKKSSNRLTG CPRGSKARDV 180 YYATFFLARP PPAAPPTGCS SLPMLEPKWP GAYPYDGSRS GGYGFTEEWL AGSDAIVNDG 240 SSAFFSEEHV SNLNFGWCSS GAEGFDLQSY CKK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 109 | 117 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
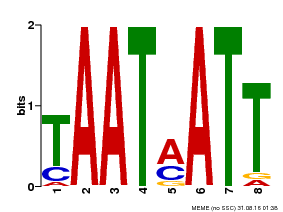
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353965 | 0.0 | AK353965.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1004B20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020190062.1 | 1e-143 | homeobox-leucine zipper protein HOX13-like | ||||
| Swissprot | Q10QP3 | 7e-84 | HOX13_ORYSJ; Homeobox-leucine zipper protein HOX13 | ||||
| TrEMBL | M8AYN4 | 0.0 | M8AYN4_AEGTA; Homeobox-leucine zipper protein HOX13 | ||||
| STRING | EMT09592 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4283 | 31 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65310.2 | 6e-29 | homeobox protein 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



