 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT14735 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 612aa MW: 68385.9 Da PI: 6.2928 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 418.6 | 6.8e-128 | 35 | 418 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX...XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqll.edkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekve 100
eel +rmwkd+++l+r+ker+++l +++e +++++k +++++qa rkkm+ra DgiLkYMlk mevcna+GfvYgiipekgkpv+gasd++raWWkekv+
EMT14735 35 EELARRMWKDRVRLRRIKERQQRLAlQQAELELEKSKPKQISDQAMRKKMARAHDGILKYMLKLMEVCNARGFVYGIIPEKGKPVSGASDNIRAWWKEKVK 135
8*******************9996414455559999999************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GGG- CS
EIN3 101 fdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkphdl 201
fd+ngpaai+ky+a++l+ ++++s+ ++ hsl++lqD+tlgSLLs+lmqhc+ppqr++plekg++pPWWP G+e+ww++lgl++ q pykkphdl
EMT14735 136 FDKNGPAAIAKYEAEHLVDADAQSS---VVKNEHSLMDLQDATLGSLLSSLMQHCNPPQRKYPLEKGTPPPWWPAGNEEWWAALGLPRGQI-APYKKPHDL 232
******************8877777...789***********************************************************9.8******** PP
-HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX.XX......XXXXXXXXXXXXXXXXXXXXXXXXXX.XXX CS
EIN3 202 kkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah.ss......slrkqspkvtlsceqkedvegkkeskikhv 295
kk+wkv+vLt+vikhmsp++++ir+++r+sk lqdkm+akes ++l vl++ee++++ + s+ ++ ++ +s+++++dv+g +e + +
EMT14735 233 KKVWKVGVLTCVIKHMSPNFDKIRNHVRKSKILQDKMTAKESLIWLGVLQREERLVHGIDNGvSEithrsaPEDRNGTRNAHSSSNEYDVDGFEEAPLSIS 333
********************************************************999888543555544334477788899999*****8888777777 PP
XXXXXXX..................................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 296 qavktta..................................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisq 351
++++++ + + krk+ +++s +v ++ev q+ +++ + ++d+n+++q
EMT14735 334 SKDDEQGlspaaqsseehvsrrdrerantkhlyqavvlkegTKKQPPKRKRARHSSVAV-EQEV----QRTDDAPENPGNLIPDMNRLDQ 418
77776558888888777777777777777777777755553333333333333333332.2223....3333333455666666666666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 3.2E-123 | 35 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.4E-67 | 159 | 291 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 8.89E-58 | 165 | 287 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000165 | Biological Process | MAPK cascade | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006612 | Biological Process | protein targeting to membrane | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0009743 | Biological Process | response to carbohydrate | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0010363 | Biological Process | regulation of plant-type hypersensitive response | ||||
| GO:0031348 | Biological Process | negative regulation of defense response | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0043069 | Biological Process | negative regulation of programmed cell death | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 612 aa Download sequence Send to blast |
MDQLALLATE FGDSSDFEVD GINENDVSDE EIEPEELARR MWKDRVRLRR IKERQQRLAL 60 QQAELELEKS KPKQISDQAM RKKMARAHDG ILKYMLKLME VCNARGFVYG IIPEKGKPVS 120 GASDNIRAWW KEKVKFDKNG PAAIAKYEAE HLVDADAQSS VVKNEHSLMD LQDATLGSLL 180 SSLMQHCNPP QRKYPLEKGT PPPWWPAGNE EWWAALGLPR GQIAPYKKPH DLKKVWKVGV 240 LTCVIKHMSP NFDKIRNHVR KSKILQDKMT AKESLIWLGV LQREERLVHG IDNGVSEITH 300 RSAPEDRNGT RNAHSSSNEY DVDGFEEAPL SISSKDDEQG LSPAAQSSEE HVSRRDRERA 360 NTKHLYQAVV LKEGTKKQPP KRKRARHSSV AVEQEVQRTD DAPENPGNLI PDMNRLDQVE 420 IPGMANHQIT SFNQMAVTSE ALQHRGNAQG HVYLPGSGVN GFDNAQPVDA TPVSIYQPVL 480 YGSSDNARSK SGSPFPLHAN SGFNSFPSNY QTLPPKQSVP LPMMDHHVVP MGIRAPADNS 540 PYGDHVIGGG SSTSVPGDMQ HLVDFPFYGE QDKFVGSSFE GLPLDYISMS SPIPDIDDLL 600 LHDDDLMEYL GT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-62 | 168 | 289 | 12 | 133 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
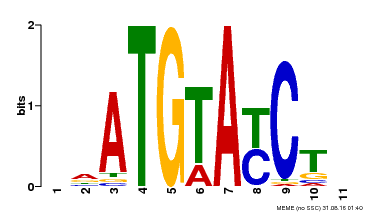
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK249582 | 0.0 | AK249582.1 Hordeum vulgare subsp. vulgare cDNA clone: FLbaf40h12, mRNA sequence. | |||
| GenBank | AK364187 | 0.0 | AK364187.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2022H23. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020187998.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-142 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A3B6MUW5 | 0.0 | A0A3B6MUW5_WHEAT; Uncharacterized protein | ||||
| TrEMBL | A0A453L4Q1 | 0.0 | A0A453L4Q1_AEGTS; Uncharacterized protein | ||||
| TrEMBL | N1QWS5 | 0.0 | N1QWS5_AEGTA; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | EMT14735 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-141 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



