 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT16558 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 267aa MW: 28382.1 Da PI: 6.885 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 49.6 | 5.4e-16 | 163 | 199 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C+nCg+ + Tp +Rrgp g++tLCnaCGl+++ kg+
EMT16558 163 CQNCGVNSrlTPAMRRGPAGPRTLCNACGLMWANKGT 199
**********************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 9.7E-9 | 30 | 65 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 15.169 | 30 | 65 | IPR010399 | Tify domain |
| Pfam | PF06200 | 1.9E-11 | 31 | 63 | IPR010399 | Tify domain |
| PROSITE profile | PS51017 | 13.354 | 92 | 134 | IPR010402 | CCT domain |
| Pfam | PF06203 | 1.0E-15 | 92 | 132 | IPR010402 | CCT domain |
| SMART | SM00401 | 1.2E-10 | 157 | 211 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 1.81E-10 | 161 | 210 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 1.4E-13 | 162 | 201 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 9.84E-15 | 163 | 209 | No hit | No description |
| Pfam | PF00320 | 1.7E-13 | 163 | 199 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 163 | 190 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 267 aa Download sequence Send to blast |
MAADGQDPPL ADAAAAASGD DDAAAADALL SPASEQLTLL YQGEVYVFDP VPPQKVQAAL 60 LVLGGCEVPP GLVSMAGPTA YGEKSTTVAA KRVASLMRFR EKRKERCFDK KIRYGVRKEV 120 AQKMKRRKGQ FAGRADFGDA ASSSAACVSA ADGEDDHFRE SHCQNCGVNS RLTPAMRRGP 180 AGPRTLCNAC GLMWANKGTL RSPLNAPKMT VQHPANLSKM ESVDDDKAIV CAEPNHTTVK 240 MDSGMSPEQE QKPELRPATE GDSMADS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250|UniProtKB:Q8LAU9}. | |||||
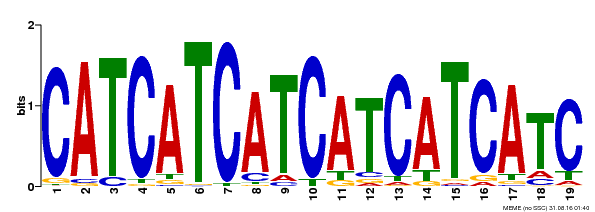
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00198 | DAP | Transfer from AT1G51600 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA), and drought and salt stresses. Down-regulated by jasmonate and wounding. {ECO:0000269|PubMed:19618278}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK376700 | 0.0 | AK376700.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3132B09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020187640.1 | 0.0 | GATA transcription factor 19-like | ||||
| Swissprot | Q0DNU1 | 1e-136 | GAT19_ORYSJ; GATA transcription factor 19 | ||||
| TrEMBL | N1R1N8 | 0.0 | N1R1N8_AEGTA; GATA transcription factor 25 | ||||
| STRING | EMT16558 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3460 | 37 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51600.2 | 1e-51 | ZIM-LIKE 2 | ||||



