 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT25826 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 517aa MW: 58101.8 Da PI: 5.01 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 108.1 | 6.5e-34 | 98 | 189 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellekik 98
Fl k+ye++ed+++++++sw g sfvv+++ +f++++LpkyFkh+nf+SF+RQLn+YgF+k++ e+ weF+++ F +g+ +ll++i+
EMT25826 98 FLIKTYEMVEDPATNRVVSWGPGGASFVVWNPPDFSRDLLPKYFKHNNFSSFIRQLNTYGFRKIDPER---------WEFANDDFIRGHMHLLKNIH 185
9****************************************************************999.........******************** PP
XXXX CS
HSF_DNA-bind 99 rkks 102
r+k
EMT25826 186 RRKP 189
*985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00415 | 4.8E-61 | 94 | 187 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 4.49E-35 | 94 | 187 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.1E-37 | 95 | 187 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00447 | 2.9E-30 | 98 | 187 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.2E-19 | 98 | 121 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.2E-19 | 136 | 148 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 137 | 161 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.2E-19 | 149 | 161 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 517 aa Download sequence Send to blast |
MGRRLGTRSP QADFSDGSPP EQVDSIDDKS LSLPTWLQRI FHRVRLDSCE QRAAPNRIQS 60 NPVEDFLAEL GRSTGMDIDM EGGSQPQPQG ASLSPAPFLI KTYEMVEDPA TNRVVSWGPG 120 GASFVVWNPP DFSRDLLPKY FKHNNFSSFI RQLNTYGFRK IDPERWEFAN DDFIRGHMHL 180 LKNIHRRKPV HSHSPQTQVN GPLAEAERRE YEEDISRLKH ENSVLVAELQ RQARQQCGLS 240 WLMQSLEDRL MAMERRQADV VASVRDTLQR RRGGVHPGQQ TMLELEPTDH FSKKRRVPRL 300 GFFVEEHSAA AAEEQRVPHL RAMGGETPGM VMVNAEPFEK MELALVSMEK LVQRAGDYAS 360 SEDMYNAAAA APSTDDPAHA DLQAAPVNLQ PSSPEIAESP GYAVQSPMLL FPEIHEDKHK 420 TMAEVDMSSE ASTTDTSQDE TTAEAEAETG VPHEPAVAND LFWERFLTDT PKPLAVEDSH 480 ESKDDVKTGL DCYWFGHRNN VEQITEQMGH LASAQKT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 2e-23 | 75 | 187 | 11 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 2e-23 | 75 | 187 | 11 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 2e-23 | 75 | 187 | 11 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
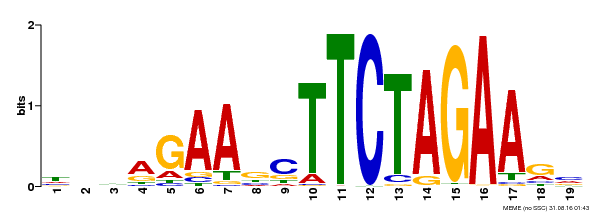
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat stress. {ECO:0000269|PubMed:12032317}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF208547 | 0.0 | KF208547.1 Triticum aestivum heat shock factor A4e (HsfA4e) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020168481.1 | 0.0 | heat stress transcription factor A-4d-like | ||||
| Swissprot | Q93VB5 | 1e-155 | HFA4D_ORYSJ; Heat stress transcription factor A-4d | ||||
| TrEMBL | M8CQB9 | 0.0 | M8CQB9_AEGTA; Heat stress transcription factor A-4d | ||||
| STRING | EMT25826 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3296 | 37 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 3e-66 | heat shock transcription factor A4A | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



