 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT26237 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 898aa MW: 98228.5 Da PI: 6.623 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.3 | 5.8e-35 | 191 | 266 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+Cqv+gCead+ e+k yh+rh+vC ++++++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
EMT26237 191 RCQVPGCEADIRELKGYHKRHRVCLRCAHSSAVMIDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 266
6************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.4E-27 | 184 | 253 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.419 | 189 | 266 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.14E-33 | 190 | 269 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.2E-26 | 192 | 266 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 898 aa Download sequence Send to blast |
MASAHLALRR SLSYNGSNRT GGVDAAGAGG GGAAADAGEP VWDWDNLLDF TIPEDDSLLL 60 PWDDPLGIEG DPTNEGALLP APPPPQPVEA EPAPPPPSPP AEAGGSRRGV RKRDPRLVCP 120 NYLAGIVPCA CPELDEMAAA AEVEEVAAEA LAGPRKKPRA ASRGSSGAVV KIGGAGGGSG 180 VAGRGGAAEM RCQVPGCEAD IRELKGYHKR HRVCLRCAHS SAVMIDGVQK RYCQQCGKFH 240 ILLDFDEDKR SCRRKLERHN KRRRRKPDSK GTLEKEIDEQ LDLSADGSGN GEIREENIDG 300 ASCDMLETVL SNKVLDRETP VGSEDALSSP TCTQLSLQND RSKSIVTFAA SAEACLGAKQ 360 ENAKLTNSPV HDSKSAYSSS CPTGRISFKL YDWNPAEFPR RLRNQIFEWL SSMPVELEGY 420 IRPGCTILTV FIAMPQHMWD KLAEDTANLV RNLVNAPSSL LLGKGAFFVH ANNTIFQVLK 480 DGATLMSTRL EVQAPRIHCV HPTWFEAGKP IELLLCGSSL DQPKFRSLLS FDGEYLKHDC 540 CRLTSRGTFG CVKNGAPTFD SHHEVFRINI TQTKPDTHGP GFVEGTKYVH ETLQADKRHR 600 QVENVFGLSN FVPILFGSKQ LCFELERIQD ALCGSSKYKS ANGEFPGITS DPCEHWKLQQ 660 TAMSGFLIEI GWLIKKPSPD EFKNLLSKTN VKRWICVLEF LIQNNFINVL EMIVKSSDNI 720 IGSEILSNLE SGRLEHHVTA FLGYIRHARI IVDQRAKHNE ETQLQTRWCG DSVSDQPSLG 780 TSVPLGKENV TASDDFCLPS TNAECEEEES VPLVTNEAVS HRQCCPSEMN ARWLNPALVA 840 PFPSGVMRTR LVATVAVAAI LCFTACVAVF HPDRVGVLAA PVKRYLFRDC PPWHHSGC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 2e-37 | 191 | 273 | 5 | 87 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 248 | 266 | KRSCRRKLERHNKRRRRKP |
| 2 | 252 | 264 | RRKLERHNKRRRR |
| 3 | 259 | 266 | NKRRRRKP |
| 4 | 260 | 265 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
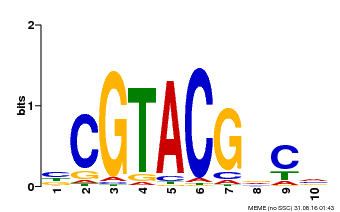
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK250312 | 0.0 | AK250312.1 Hordeum vulgare subsp. vulgare cDNA clone: FLbaf72c21, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020161545.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | M8CGD6 | 0.0 | M8CGD6_AEGTA; Squamosa promoter-binding-like protein 9 | ||||
| STRING | EMT26237 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-137 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



