 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT29878 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 355aa MW: 37495.3 Da PI: 8.0398 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 185.8 | 1.7e-57 | 24 | 161 | 12 | 147 |
DUF822 12 rEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl.eeaeaagssasaspesslqsslkssalaspvesys 111
rEnn+rRERrRRaiaakiy+GLRa+Gny+lpk++DnneVlkALc+eAGwvve+DGttyr+g+kp+ ++ +sasasp+ss+q +sp++sy+
EMT29878 24 RENNRRRERRRRAIAAKIYTGLRAYGNYNLPKHCDNNEVLKALCNEAGWVVEPDGTTYRRGCKPPpQARTDPMRSASASPCSSYQ--------PSPRASYN 116
9****************************************************************88889999************........******** PP
DUF822 112 aspksssfpspssldsislasa.........asllpvlsvlslvs 147
+sp+sssfps+ s+++i+l +sl+p+l++ls+ +
EMT29878 117 PSPASSSFPSSGSSSHITLGGGnnfiggvegSSLIPWLKNLSSNP 161
*****************9987678999999999999999998744 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.7E-62 | 24 | 158 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 355 aa Download sequence Send to blast |
MADPAAGAGG AERERRAGGR HGLRENNRRR ERRRRAIAAK IYTGLRAYGN YNLPKHCDNN 60 EVLKALCNEA GWVVEPDGTT YRRGCKPPPQ ARTDPMRSAS ASPCSSYQPS PRASYNPSPA 120 SSSFPSSGSS SHITLGGGNN FIGGVEGSSL IPWLKNLSSN PSFASSSKLP QLHHLYFNGG 180 SISAPVTPPS SSPTHTPRMK TDWESQCVLP PWAGANYASL PNSTPPSPGH HVAPDPAWLA 240 GFQISSAGPS SPTYNLVSHN PFGIALASSS RVCTPGQSGT CSPAMGDHAP AHHDVQMEMV 300 EGAADDFAFG SNSNGNNGSP GLVKAWEGER IHEECASDEL ELTLGSSRTR GEPPF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 4e-25 | 36 | 89 | 391 | 444 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 4e-25 | 36 | 89 | 391 | 444 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 4e-25 | 36 | 89 | 391 | 444 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 4e-25 | 36 | 89 | 391 | 444 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May function in brassinosteroid signaling. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
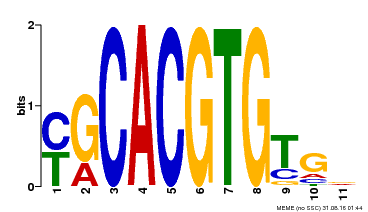
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK360181 | 0.0 | AK360181.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1112F17. | |||
| GenBank | AK366794 | 0.0 | AK366794.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2046O02. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020152195.1 | 0.0 | protein BZR1 homolog 3-like | ||||
| Swissprot | Q5Z9E5 | 1e-173 | BZR3_ORYSJ; Protein BZR1 homolog 3 | ||||
| TrEMBL | M8C751 | 0.0 | M8C751_AEGTA; Uncharacterized protein | ||||
| STRING | EMT29878 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1366 | 38 | 112 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 4e-68 | BES1/BZR1 homolog 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



