 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EMT31332 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Aegilops
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 333aa MW: 36415.1 Da PI: 10.1442 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 31.3 | 4.9e-10 | 60 | 106 | 1 | 53 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkl 53
s++kGV + +grW ++I++ r +r++lg+f + Aa+a+++a +++
EMT31332 60 SRFKGVVPQP-NGRWGSQIYE------RhARVWLGTFADQDLAARAYDEAELAF 106
689***9788.8********9......44*********************8875 PP
| |||||||
| 2 | B3 | 94.4 | 7.8e-30 | 154 | 249 | 1 | 87 |
EEEE-..-HHHHTT-EE--HHH.HTT.........---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.........ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+ tpsdv+k++rlv+pk++ae+h + +++ l +ed +g++W+++++y+++s++yvltkGW++Fv+++gL +gD++vF+ +
EMT31332 154 FEKAVTPSDVGKLNRLVVPKQHAEKHfplkrtpetTTTTGNGVLLNFEDGEGKVWRFRYSYWNSSQSYVLTKGWSRFVREKGLAAGDSIVFSCSAY 249
89*************************99877766444458999***********************************************96543 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 2.88E-10 | 60 | 107 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 3.7E-5 | 60 | 105 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.33E-15 | 60 | 106 | No hit | No description |
| SMART | SM00380 | 8.4E-11 | 61 | 114 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.079 | 61 | 119 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.4E-13 | 61 | 110 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 2.6E-37 | 149 | 264 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 4.05E-29 | 151 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.27E-28 | 152 | 244 | No hit | No description |
| SMART | SM01019 | 5.0E-23 | 154 | 262 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.662 | 154 | 263 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.5E-27 | 154 | 251 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MGVEILSSTV EHYSQYSSSA STATTESGAA GRSPMALSLP VAIADESVTS RSTSAQLALS 60 RFKGVVPQPN GRWGSQIYER HARVWLGTFA DQDLAARAYD EAELAFLAAH SKAEIVDMLR 120 KHTYADELRQ GLRRGRGMGT RAQPTPSWAR EPLFEKAVTP SDVGKLNRLV VPKQHAEKHF 180 PLKRTPETTT TTGNGVLLNF EDGEGKVWRF RYSYWNSSQS YVLTKGWSRF VREKGLAAGD 240 SIVFSCSAYG QEKQFFINCK KNTTVNGGKS ASPLPVVETA KGEQVRVVRL FGVDIAGVKR 300 GRAATAEHGP PELFKRQCVA HGQHSPALGA FVL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 4e-47 | 151 | 260 | 11 | 115 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
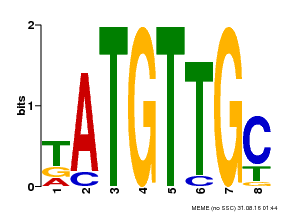
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020191173.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Refseq | XP_020200371.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Refseq | XP_020201146.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0141000-like isoform X1 | ||||
| Swissprot | Q9AWS0 | 1e-128 | Y1410_ORYSJ; AP2/ERF and B3 domain-containing protein Os01g0141000 | ||||
| TrEMBL | M8CUQ6 | 0.0 | M8CUQ6_AEGTA; AP2/ERF and B3 domain-containing protein | ||||
| STRING | EMT31332 | 0.0 | (Aegilops tauschii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 2e-89 | related to ABI3/VP1 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



