 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EPS59646.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Genlisea
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 178aa MW: 20276.3 Da PI: 10.7179 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 92.8 | 2.9e-29 | 21 | 75 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
kp+++WtpeLH+rFv+aveqL G +kA+P++ilelm +++Lt+++v+SHLQkYR++
EPS59646.1 21 KPKVDWTPELHRRFVQAVEQL-GVDKAVPSRILELMRIDCLTRHNVASHLQKYRSH 75
79*******************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.716 | 18 | 77 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-28 | 19 | 78 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.27E-20 | 19 | 78 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.4E-27 | 21 | 75 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.6E-8 | 24 | 73 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 178 aa Download sequence Send to blast |
KDENKPKKSI PSSKNATGKK KPKVDWTPEL HRRFVQAVEQ LGVDKAVPSR ILELMRIDCL 60 TRHNVASHLQ KYRSHRKHLL AREAVAASWN QRRKIISSGK KDHVSPWITP PPTTTIGFPP 120 MPPLHVWGHP SLDQSLMHTW PKHNHPPPSP PSWPPTPVDP QLEFFYAPIS NCDSNFSP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-19 | 17 | 73 | 1 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that functions with GLK2 to promote chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. Acts in a cell-autonomous manner to coordinate and maintain the photosynthetic apparatus within individual cells. May function in photosynthetic capacity optimization by integrating responses to variable environmental and endogenous cues (PubMed:11828027, PubMed:12220263, PubMed:17533111, PubMed:18643989, PubMed:19376934, PubMed:19383092, PubMed:19726569). Prevents premature senescence (PubMed:23459204). {ECO:0000269|PubMed:11828027, ECO:0000269|PubMed:12220263, ECO:0000269|PubMed:17533111, ECO:0000269|PubMed:18643989, ECO:0000269|PubMed:19376934, ECO:0000269|PubMed:19383092, ECO:0000269|PubMed:19726569, ECO:0000269|PubMed:23459204}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
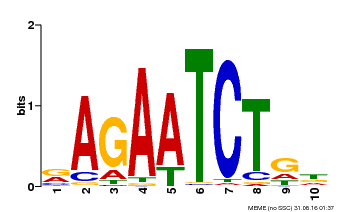
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. Repressed by BZR2. {ECO:0000269|PubMed:12220263, ECO:0000269|PubMed:21214652}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021279775.1 | 5e-66 | transcription activator GLK1 | ||||
| Swissprot | Q9SIV3 | 1e-52 | GLK1_ARATH; Transcription activator GLK1 | ||||
| TrEMBL | S8C5C7 | 1e-126 | S8C5C7_9LAMI; Uncharacterized protein (Fragment) | ||||
| STRING | Migut.M00147.1.p | 3e-63 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2532 | 23 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 3e-44 | GBF's pro-rich region-interacting factor 1 | ||||



