 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EPS62844.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Genlisea
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 480aa MW: 53526.8 Da PI: 7.455 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2e-23 | 134 | 235 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefel 92
f+k+lt sd++++g +++p++ ae+ + + ++l+ +d++ +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F ++++ +l
EPS62844.1 134 FCKTLTASDTSTHGGFSVPRRAAEKVfppldfsQT--PPAQELIARDLHDVEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI--WNDKNQL 228
99*****************************7444..44459************************************************..7888889 PP
EEEEE-S CS
B3 93 vvkvfrk 99
+++++r+
EPS62844.1 229 LLGIRRA 235
****997 PP
| |||||||
| 2 | Auxin_resp | 120.4 | 1.4e-39 | 260 | 343 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v+YnPras+seFv++++k+ ka++ ++vsvGmRf+m fete+ss rr++Gt++g+ +ldpvrWpnS+Wrs+k
EPS62844.1 260 AAHAAATNSCFTVFYNPRASPSEFVIPLSKYAKAVYhTRVSVGMRFRMLFETEESSVRRYMGTITGIGELDPVRWPNSHWRSVK 343
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 8.37E-49 | 118 | 263 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.1E-41 | 127 | 249 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 8.11E-22 | 133 | 234 | No hit | No description |
| Pfam | PF02362 | 1.4E-21 | 134 | 235 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.2E-24 | 134 | 236 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.323 | 134 | 236 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 6.6E-35 | 260 | 343 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 480 aa Download sequence Send to blast |
VERIDESMKL STSATSQQVH EGEKKCLNSE LWHACAGPLV SLPSVGSRVV YFPQGHSEQV 60 AATTNREVDA QLPNYPTLPP QLICQLHNVT MHADVETDEV YAQMTLQPLA PVEEKDTYLP 120 VEFGIPSRQP TNYFCKTLTA SDTSTHGGFS VPRRAAEKVF PPLDFSQTPP AQELIARDLH 180 DVEWKFRHIF RGQPKRHLLT TGWSVFVSAK RLVAGDSVLF IWNDKNQLLL GIRRASRPQT 240 VMPSSVLSSD SMHIGLLAAA AHAAATNSCF TVFYNPRASP SEFVIPLSKY AKAVYHTRVS 300 VGMRFRMLFE TEESSVRRYM GTITGIGELD PVRWPNSHWR SVKVGWDEST AGDRQPRVSL 360 WEIEPLTTFP MYPSLFPLRL KRPWYPGASS FQDAGNEAMN SSMAWLRGES GEPGGFNALN 420 FQSVGMFPVV QQRFDPTAMR NDVGQYQAML AAAGMHGFGS GSDLMKHQVM QFQQLTSVNS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-165 | 24 | 364 | 48 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF6. Involved in fruit initiation. Acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
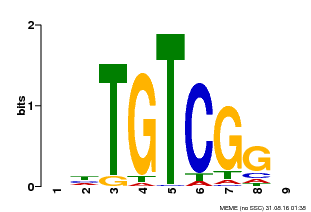
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027075605.1 | 0.0 | auxin response factor 8-like | ||||
| Swissprot | Q9FGV1 | 0.0 | ARFH_ARATH; Auxin response factor 8 | ||||
| TrEMBL | S8C7S5 | 0.0 | S8C7S5_9LAMI; Auxin response factor (Fragment) | ||||
| STRING | XP_009789648.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3857 | 23 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.2 | 0.0 | auxin response factor 8 | ||||



