 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EPS63500.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Genlisea
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 185aa MW: 21046.3 Da PI: 10.3006 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 177.4 | 3.9e-55 | 11 | 136 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgel 99
lppGfrFhPtdeel+++yL +k+ + +++ ++ +vd++kvePwdLp k+k +ekewyfF+ rd+ky+tg r+nrat +gyWkatgkdke+l+ +g l
EPS63500.1 11 LPPGFRFHPTDEELITHYLSPKIINAGFTT-GIMGDVDLNKVEPWDLPWKAKMGEKEWYFFCIRDRKYPTGLRTNRATGAGYWKATGKDKEILR-GGIL 107
79*************************999.89***************888999***************************************9.**** PP
NAM 100 vglkktLvfykgrapkgektdWvmheyrl 128
vg+kktLvfykgrapkgekt+W+mheyr
EPS63500.1 108 VGMKKTLVFYKGRAPKGEKTNWIMHEYRQ 136
***************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.28E-64 | 7 | 159 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.378 | 11 | 159 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.1E-29 | 12 | 135 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 185 aa Download sequence Send to blast |
GMAGGDDQFS LPPGFRFHPT DEELITHYLS PKIINAGFTT GIMGDVDLNK VEPWDLPWKA 60 KMGEKEWYFF CIRDRKYPTG LRTNRATGAG YWKATGKDKE ILRGGILVGM KKTLVFYKGR 120 APKGEKTNWI MHEYRQETKN DLFKAANKND WVISRVFKKS PGGKKVHISG LIKNQRTGDI 180 PPLID |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 7e-58 | 8 | 165 | 14 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 7e-58 | 8 | 165 | 14 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 7e-58 | 8 | 165 | 14 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 7e-58 | 8 | 165 | 14 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 9e-58 | 8 | 165 | 17 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 7e-58 | 8 | 165 | 14 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 7e-58 | 8 | 165 | 14 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the promoter regions of genes involved in chlorophyll catabolic processes, such as NYC1, SGR1, SGR2 and PAO. {ECO:0000269|PubMed:27021284}. | |||||
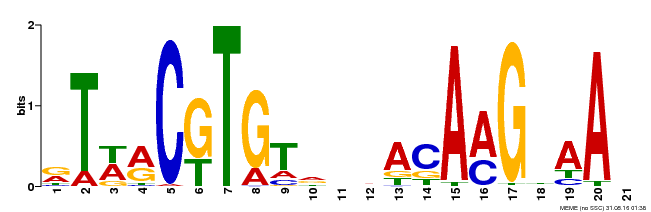
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00496 | DAP | Transfer from AT5G07680 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the microRNA miR164. {ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:17098808}. | |||||
| UniProt | INDUCTION: Repressed post-transcriptionally by miR164. {ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:17098808, ECO:0000269|PubMed:18305205, ECO:0000305|PubMed:15723790}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009761662.1 | 3e-97 | PREDICTED: NAC domain-containing protein 100-like | ||||
| Refseq | XP_016437736.1 | 3e-97 | PREDICTED: NAC domain-containing protein 92-like | ||||
| Swissprot | Q9FLJ2 | 9e-89 | NC100_ARATH; NAC domain-containing protein 100 | ||||
| Swissprot | Q9FLR3 | 9e-89 | NAC79_ARATH; NAC domain-containing protein 79 | ||||
| TrEMBL | S8C9M3 | 1e-134 | S8C9M3_9LAMI; Uncharacterized protein (Fragment) | ||||
| STRING | XP_009761662.1 | 1e-96 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA152 | 24 | 279 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G07680.1 | 4e-91 | NAC domain containing protein 80 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



