 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EPS63592.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Genlisea
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 184aa MW: 21721.7 Da PI: 9.6459 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 161 | 4.5e-50 | 15 | 140 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelv 100
+pGfrFhPt+eelv +yL++kvegk++++ e i+ +d+y+++Pw+Lp+ ++ +ekew+f+++rdkky++g+r+nr+t+sgyWkatg d+ + s++ + +
EPS63592.1 15 MPGFRFHPTEEELVEFYLRRKVEGKRFNV-ELITFLDLYRYDPWELPALAAIGEKEWFFYVPRDKKYRNGDRPNRVTTSGYWKATGADRMIRSENFQSI 112
79***************************.89***************8778899********************************************* PP
NAM 101 glkktLvfykgrapkgektdWvmheyrl 128
glkktLvfy+g+apkg +t W+m+eyrl
EPS63592.1 113 GLKKTLVFYSGKAPKGIRTCWIMNEYRL 140
**************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 54.489 | 14 | 161 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 1.44E-55 | 14 | 160 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.1E-25 | 16 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 184 aa Download sequence Send to blast |
ESHPDDDFHQ HDLVMPGFRF HPTEEELVEF YLRRKVEGKR FNVELITFLD LYRYDPWELP 60 ALAAIGEKEW FFYVPRDKKY RNGDRPNRVT TSGYWKATGA DRMIRSENFQ SIGLKKTLVF 120 YSGKAPKGIR TCWIMNEYRL PLHETERLQK GEISLCRVYK RAGVEDRPSL PRSIPSSRSS 180 KSNS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-55 | 16 | 161 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-55 | 16 | 161 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-55 | 16 | 161 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-55 | 16 | 161 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 4e-55 | 16 | 161 | 22 | 168 | NAC domain-containing protein 19 |
| 4dul_A | 4e-55 | 16 | 161 | 19 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 4e-55 | 16 | 161 | 19 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as a floral repressor. Controls flowering time by negatively regulating CONSTANS (CO) expression in a GIGANTEA (GI)-independent manner. Regulates the plant cold response by positive regulation of the cold response genes COR15A and KIN1. May coordinate cold response and flowering time. {ECO:0000269|PubMed:17653269}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00257 | DAP | Transfer from AT2G02450 | Download |
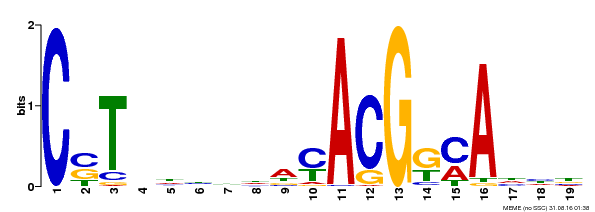
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian regulation with a peak of expression at dawn under continuous light conditions (PubMed:17653269). Circadian regulation with a peak of expression around dusk and lowest expression around dawn under continuous light conditions (at protein level) (PubMed:17653269). {ECO:0000269|PubMed:17653269}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004301581.1 | 1e-118 | PREDICTED: NAC domain-containing protein 48 | ||||
| Refseq | XP_009626107.1 | 1e-118 | PREDICTED: NAC domain-containing protein 35-like isoform X1 | ||||
| Swissprot | Q9ZVP8 | 1e-106 | NAC35_ARATH; NAC domain-containing protein 35 | ||||
| TrEMBL | S8DUK5 | 1e-134 | S8DUK5_9LAMI; Uncharacterized protein (Fragment) | ||||
| STRING | XP_009626107.1 | 1e-118 | (Nicotiana tomentosiformis) | ||||
| STRING | XP_004301581.1 | 1e-117 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3888 | 24 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02450.2 | 1e-103 | NAC domain containing protein 35 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



