| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
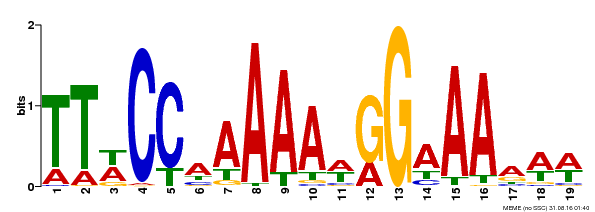
| 1 | SRF-TF | 99.7 | 1.2e-31 | 9 | 58 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
k+ienk+nrqvtfskRrng+lKKA+ELSvLCdaeva+iifss+gklye+
EPS71714.1 9 KKIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFG 58
68**********************************************95 PP
|
| 2 | K-box | 98.5 | 9.5e-33 | 78 | 169 | 7 | 98 |
K-box 7 ksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkl 98
+ ++e+++ s+qqe+akLk ++++Lqr qRhllGedL++Ls+keL++Le+qLe +l + R +K+++l+eq+e+l++ke+el + nk+L+ k+
EPS71714.1 78 ETTSERETRSWQQEIAKLKTKYDSLQRAQRHLLGEDLGPLSVKELHNLEKQLEAALLQARHRKTQMLMEQMEHLRRKERELGDMNKNLKIKI 169
33689999********************************************************************************9987 PP
|
| Protein Features
? help Back to Top |
 |
| Database |
Entry ID |
E-value |
Start |
End |
InterPro ID |
Description |
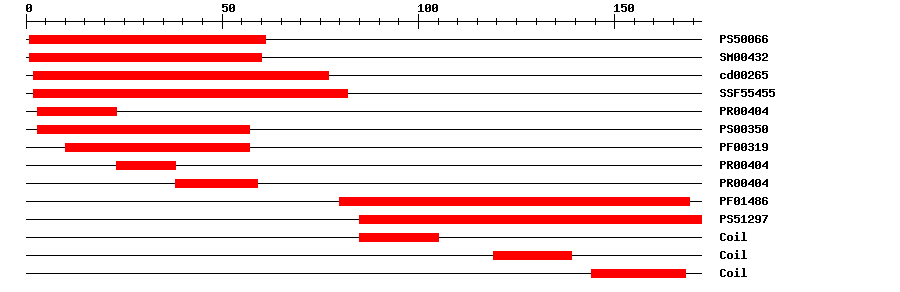
| PROSITE profile | PS50066 | 33.487 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 9.8E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 7.26E-43 | 2 | 77 | No hit | No description |
| SuperFamily | SSF55455 | 7.46E-34 | 2 | 82 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.8E-27 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 3.7E-29 | 80 | 169 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.96 | 85 | 172 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top |
| GO Term |
GO Category |
GO Description |
| GO:0009553 | Biological Process | embryo sac development |
| GO:0009911 | Biological Process | positive regulation of flower development |
| GO:0010094 | Biological Process | specification of carpel identity |
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem |
| GO:0010582 | Biological Process | floral meristem determinacy |
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated |
| GO:0048455 | Biological Process | stamen formation |
| GO:0048459 | Biological Process | floral whorl structural organization |
| GO:0048509 | Biological Process | regulation of meristem development |
| GO:0048833 | Biological Process | specification of floral organ number |
| GO:0080060 | Biological Process | integument development |
| GO:0080112 | Biological Process | seed growth |
| GO:0005634 | Cellular Component | nucleus |
| GO:0003677 | Molecular Function | DNA binding |
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding |
| GO:0046983 | Molecular Function | protein dimerization activity |