 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EPS72406.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Genlisea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 878aa MW: 97033.6 Da PI: 4.8534 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52 | 1.6e-16 | 11 | 57 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l av+++ g++Wk+Ia+++ Rt+ qc +rwqk+l
EPS72406.1 11 KGQWTQEEDEVLRIAVQRFKGKNWKKIAECFK-DRTDVQCLHRWQKVL 57
799****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.2 | 1e-19 | 63 | 109 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W+++Ede+++++vk++G + W+tI+++++ gR +kqc++rw+++l
EPS72406.1 63 KGPWSKQEDEIIIQLVKKYGAKKWSTISQHLP-GRIGKQCRERWHNHL 109
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 42.3 | 1.7e-13 | 115 | 157 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+ E++ l++a++ +G++ W+ + ++ gR+++ +k++w+
EPS72406.1 115 KEAWTQNEELMLIRAHQIYGNK-WAELTKFLP-GRSDNAIKNHWN 157
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.312 | 6 | 57 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.79E-15 | 7 | 64 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-12 | 10 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.6E-14 | 11 | 57 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.1E-23 | 13 | 68 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.54E-12 | 14 | 57 | No hit | No description |
| PROSITE profile | PS51294 | 32.609 | 58 | 113 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.68E-30 | 60 | 156 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.6E-19 | 62 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.57E-16 | 65 | 109 | No hit | No description |
| Pfam | PF13921 | 3.7E-18 | 66 | 124 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-27 | 69 | 112 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-20 | 113 | 164 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-12 | 114 | 162 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 17.162 | 114 | 164 | IPR017930 | Myb domain |
| CDD | cd00167 | 1.05E-9 | 117 | 157 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 878 aa Download sequence Send to blast |
RTSGPTRRST KGQWTQEEDE VLRIAVQRFK GKNWKKIAEC FKDRTDVQCL HRWQKVLNPE 60 LVKGPWSKQE DEIIIQLVKK YGAKKWSTIS QHLPGRIGKQ CRERWHNHLN PNINKEAWTQ 120 NEELMLIRAH QIYGNKWAEL TKFLPGRSDN AIKNHWNSSV KKKLEMYLAS GLLSQFEGSP 180 LRLPNQSMAS SSSKAQQCSE ADDFVGVDTA EMSECSQGSA AVSAPQPDMT GITHNREECG 240 ATENSSSVLC AEDYYPAFEE ANFGIPEDQS GMGEQCREND LCLDLDALSE ADWHIDPNKL 300 PEISLIDLGE EIPGLPLTSL SGPVDSSAAF DSSSSFDDMV VGDAPGLVAD LEDRMLYSEV 360 NRNEPGTSKT VNPEYGGQGD RLLHCPTFQI AAPESCCMPL DMLESALSES PPSSALRPPA 420 ETPSIFTEKL KQLSCSLQGN AGVYVSPRRR DDFVYSKESD SSPCADEVVF EGKESPRLVP 480 ANDFVLEQLN ETHSSSPFEE NFTAADKQAP GTLYYEPPRF PSLDIPFFSC DLIQSGSEIQ 540 QEYSPFGIRQ LMMSSVTPFK LWDSPSRGDD ASPEAVLKSA AKTFSGTPSI LRKRHRDLMS 600 PLSEKRDEKK PSNQDSFSSL ADDFSKLDML LDGFADAKGL PMPVSPKIKN LEVPCAEKEN 660 GGCIKTTGAS KSIESNMLSD STDASKTTEK DLGGVLVEHN INDLHFLSPH EFGSKAFGRR 720 LDSKQQHVLP SSSENSFVSP RLGAKRDCSS GSKPIAASSS ICRNSRCVDT AFKRRLESPS 780 AWKSPWFLNT DITIEDFGYF VSPGECSYDA IGLMKQLGEH TAATFADAQE VLGDETPETI 840 MKKAASSPSP SSNFFKRCIL VWLQVERRVL DFSECGSP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-67 | 11 | 164 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-67 | 11 | 164 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
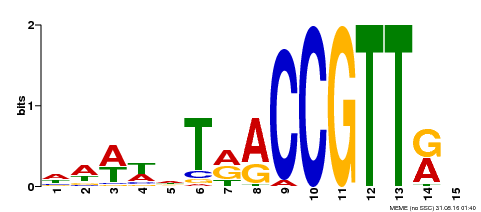
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011089667.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_011089827.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_011089990.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_020553179.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| Refseq | XP_020553183.1 | 0.0 | transcription factor MYB3R-1 isoform X1 | ||||
| TrEMBL | S8D4X3 | 0.0 | S8D4X3_9LAMI; Uncharacterized protein (Fragment) | ||||
| STRING | Migut.E01513.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3107 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 1e-159 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



