 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC027177.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 276aa MW: 31859.1 Da PI: 4.6887 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.1 | 1.7e-19 | 43 | 96 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t eq++ Le+ Fe ++++ +++ +LAk lgL+ rqV +WFqNrRa++k
EcC027177.10 43 KKRRLTVEQVQFLEKSFEVENKLEPDRKIQLAKDLGLQPRQVAIWFQNRRARWK 96
56789************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 127.5 | 5.7e-41 | 42 | 132 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv++LE+sFe e+kLep+rK +la++Lglqprqva+WFqnrRAR+ktkqlEkdye+L++++++lk++ ++L ke ++L++e+
EcC027177.10 42 EKKRRLTVEQVQFLEKSFEVENKLEPDRKIQLAKDLGLQPRQVAIWFQNRRARWKTKQLEKDYETLQASFNTLKSDYDTLIKERNDLKAEV 132
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.71E-20 | 30 | 100 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.329 | 38 | 98 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-19 | 41 | 102 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 9.1E-17 | 43 | 96 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.28E-17 | 43 | 99 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-20 | 45 | 105 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.0E-5 | 69 | 78 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 73 | 96 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.0E-5 | 78 | 94 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.3E-14 | 98 | 139 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
SRAMVSFEDA HRGNRLNRPF FRSIDPDENG EDDLEDYFHQ PEKKRRLTVE QVQFLEKSFE 60 VENKLEPDRK IQLAKDLGLQ PRQVAIWFQN RRARWKTKQL EKDYETLQAS FNTLKSDYDT 120 LIKERNDLKA EVLNLTDKLL HKGKEKESSE SSSKSSQGLF QNPVADSVSE DEVSRVPIPT 180 CRPEDICSVK SGMFDSESPH YTDAAHSSLL EPGDSSYAFE PDHSDLSQDE EDNLSKSLLS 240 TRNYPKLENS DYAILPPNTC NFGFHAEDPA FWPWSY |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 90 | 98 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
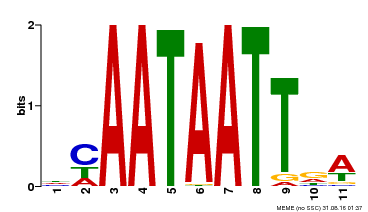
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010028877.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A059CYV1 | 0.0 | A0A059CYV1_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010028877.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM545 | 28 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 2e-45 | homeobox 1 | ||||



