 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC031757.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 659aa MW: 73803.5 Da PI: 6.605 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 39.6 | 1.4e-12 | 116 | 166 | 1 | 56 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwen 56
+Wt +evlaL ++r+ me+++ + We++s+k++e gf+r++ +Ck+k+e+
EcC031757.10 116 QWTSDEVLALFKIRSGMENWFPEF-----IWEHISRKLAELGFRRTAAECKDKFEE 166
7********************998.....9************************96 PP
| |||||||
| 2 | trihelix | 95.1 | 6.5e-30 | 503 | 598 | 1 | 86 |
trihelix 1 rWtkqevlaLiearr...........emeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++ev+aLi++r ++e + ++g+ ++plWe++s+km+e g+ers+k+Ckekwen+nk+++k+k+++kkr s +s+tcpyf+ql
EcC031757.10 503 RWPRDEVQALINLRCslyggtnnedhKLEGSNNNGNNRAPLWERISQKMSELGYERSAKRCKEKWENINKYFRKTKDNDKKR-SLDSRTCPYFHQLS 598
8**************899999988888888888899*********************************************8.9999********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 25 | 113 | 170 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 3.8E-8 | 115 | 166 | No hit | No description |
| PROSITE profile | PS50090 | 6.121 | 117 | 168 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.062 | 500 | 573 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.52E-23 | 502 | 578 | No hit | No description |
| Pfam | PF13837 | 9.0E-20 | 502 | 599 | No hit | No description |
| PROSITE profile | PS50090 | 6.679 | 503 | 571 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 659 aa Download sequence Send to blast |
MFDGVPDQFH QFLASSRASS LSLPLSFPLQ HASSPPPPSL ASLNFAAFDL YPPQHHHLQQ 60 PPPQFLQPLH QPPRAQKDII HEEIDGENSL VPVNLGIDQR ERSKLAEAAT VDDDRQWTSD 120 EVLALFKIRS GMENWFPEFI WEHISRKLAE LGFRRTAAEC KDKFEEESIS SYTPASASYP 180 KNDRLLSELE ELCNSNHGDH QNNPPILAAD KSTDNKVGKL RAGEEEDKMA GQMIQNFDGT 240 NSRNELRATV NDASEQNYKK AATDDKDHIN RSSSKGKKRK RSQEERLVKY ELFKGLCESV 300 VQRLMAQQEE MHNKLLEDMV RREKEMVEKE EAWKRQEMER MNKELDARAH EQAIAGNRQA 360 TIIEFLKKFT SFDGDDDHSP HLLSNSSSPC SLAEPNSSNP NDNSSTSHMG THKQSHAAEV 420 PTSSFLENHN HSTCSSSFQS TPTKQDPSSA SAAMAQTESR SNPPTLTAMT PQNPNPPNPH 480 QKTHHGSQSS APNAAKDQDL GKRWPRDEVQ ALINLRCSLY GGTNNEDHKL EGSNNNGNNR 540 APLWERISQK MSELGYERSA KRCKEKWENI NKYFRKTKDN DKKRSLDSRT CPYFHQLSHL 600 YSQGKLAATA TATATATVPP ADQPLPATSA HGDSSAAQLS AGRDKDVANQ VAPAANFEF |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 276 | 281 | KKRKRS |
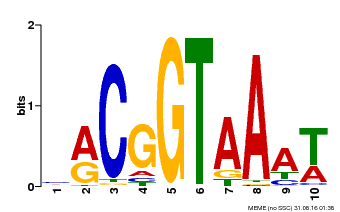
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010025058.1 | 0.0 | PREDICTED: trihelix transcription factor GTL2 | ||||
| TrEMBL | A0A059B601 | 0.0 | A0A059B601_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010025058.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8268 | 28 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 4e-66 | Trihelix family protein | ||||



