 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC033290.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 607aa MW: 65451.3 Da PI: 5.9862 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 80.5 | 1.9e-25 | 159 | 209 | 1 | 52 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQk 52
kpr++W+ eLH++Fv+av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQ+
EcC033290.10 159 KPRVVWSVELHQQFVSAVNQL-GIDKAVPKRILELMNVPGLTRENVASHLQR 209
79*******************.*****************************7 PP
| |||||||
| 2 | Response_reg | 55.4 | 3.5e-19 | 2 | 84 | 28 | 109 |
EESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 28 eaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGakdflsKpfdpeelvk 109
++++++ al+ll+ek+ +D++l D+ mp+mdG++ll++ e +lp+i++++ g+ + + + ++ Ga d+l Kp+ peel +
EcC033290.10 2 TCAQATVALNLLREKKgcFDIVLSDVHMPDMDGYKLLEHVGLEM-DLPVIMMSGDGSTNAVMRGIRHGACDYLIKPIRPEELQN 84
6778999*******999***********************6644.8***********************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00156 | 1.02E-19 | 1 | 89 | No hit | No description |
| PROSITE profile | PS50110 | 32.649 | 1 | 90 | IPR001789 | Signal transduction response regulator, receiver domain |
| Gene3D | G3DSA:3.40.50.2300 | 4.3E-29 | 1 | 108 | No hit | No description |
| SMART | SM00448 | 0.0035 | 1 | 86 | IPR001789 | Signal transduction response regulator, receiver domain |
| SuperFamily | SSF52172 | 1.17E-24 | 2 | 97 | IPR011006 | CheY-like superfamily |
| Pfam | PF00072 | 1.3E-15 | 3 | 84 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS51294 | 9.831 | 156 | 215 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-26 | 156 | 209 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.94E-16 | 156 | 209 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.1E-22 | 159 | 209 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.0E-6 | 161 | 209 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 607 aa Download sequence Send to blast |
TTCAQATVAL NLLREKKGCF DIVLSDVHMP DMDGYKLLEH VGLEMDLPVI MMSGDGSTNA 60 VMRGIRHGAC DYLIKPIRPE ELQNIWQHVV RKKWNGNKEI EHSGSLEDND RHKRGSDDAD 120 GSAVNEGGEG LLKNQKKRSI AKEEDEPELD SDDPTTSKKP RVVWSVELHQ QFVSAVNQLG 180 IDKAVPKRIL ELMNVPGLTR ENVASHLQRL SGVAQQQGGI SSSFCGTVES NIKLGPLGRY 240 DIQALAASGQ ISPQALAALQ SELLGRPTSN LVLPSFDQPT LLQASLQGSK CLPVEHGVAF 300 GQPLIKCPSN VSKQYQQSIL SSQDAPSGFG PWQATSLSTV GNNNIVAGSN IQSNNTLMDI 360 FQRQQQYQQK QQLQQQQQQQ QQQQQQLPQQ PLLEPSRLIN VQPSCLVVPH SSAGFQAAGS 420 PTSVNQSSTY NRSAVIDYSL PSYQGNTALN TGLASDKDLK TSGVLGGYSV QGSISPTLSS 480 SSANADSSNC CPLRNPSAKY NIIGQAPGVV SDFSNIQGSY DAKSGEAIDQ GLLKNLGFVS 540 KGSGTPSLFT VDEFGPPLKQ IYSGKVHLES SAGRVKQEPS FDFVDAKVSI PVLEQLSSND 600 LMSVFTE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-19 | 155 | 210 | 1 | 56 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
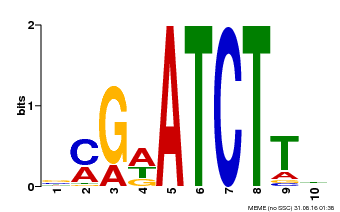
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010066564.1 | 0.0 | PREDICTED: two-component response regulator ORR21 isoform X1 | ||||
| Refseq | XP_018733087.1 | 0.0 | PREDICTED: two-component response regulator ORR21 isoform X1 | ||||
| TrEMBL | A0A059BEG2 | 0.0 | A0A059BEG2_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010066564.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8150 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 6e-95 | response regulator 2 | ||||



