 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC033786.20 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 645aa MW: 69431.4 Da PI: 6.8226 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.9 | 2.8e-16 | 433 | 479 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
EcC033786.20 433 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 479
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.03E-17 | 425 | 483 | No hit | No description |
| SuperFamily | SSF47459 | 1.02E-20 | 426 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 429 | 478 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 9.3E-14 | 433 | 479 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-20 | 433 | 487 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.6E-18 | 435 | 484 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 645 aa Download sequence Send to blast |
MAFNFVRRPA NELVELTWEH GQVVLQGQSS RAKRGPFSNN TPSNGSKPRE GASDDDTRLR 60 PGKFRNLECV LSEFPVTEQS SEIGLGQEDD MVPWLSFPVT GSLQNDYCSD LLTELTAVAA 120 NGTSPGNDDK SSSQIDRESH GIDNGVRSLG QTDASKFSSS AAGETTINRT GSNQLYSLPN 180 QQFLSPLPCF RSRALDDVST STSSSVSLAV CGDKAGLPTS VVACSTVKVQ RQQDLGHSDK 240 SSCFTNFSCF SRPASIYRVN NAESSGQTAG FGLSGVDRSV SKEKASFNDN STLSGSNFAV 300 SSDGLQKQRK IDSSKPFDRK QYEHKFSGGQ SEAICQENTC KNAKHSCQNY QASVCKGSTE 360 REKTTEHVGG SSSVCSGNSV ERVLDDPKSN LKRKSLETDE SDGPSEDIEE DPTREKKQNS 420 AQGTSKRSRA AEVHNLSERR RRDRINEKMR ALQELIPNCN KVDKASMLDE AIEYLKTLQL 480 QVQMMSMGAG LYIPPMMVPT GMHPAQMPHF PPFGIGMGSA IGFGMGMVDM SGRSSGRPVV 540 PVPPMQGPHF PSPLISAGMA GVNLQMFNFP GQGTPLPRPC APLVPFPGGP VMKPAVGLGV 600 YGMPTSTENS DSAVASRSKE AMQSIDQEPL QNTASICLKN QSSNQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 437 | 442 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
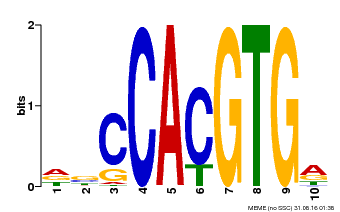
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018717209.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X2 | ||||
| TrEMBL | A0A059AXT1 | 0.0 | A0A059AXT1_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010070103.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7665 | 27 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 3e-42 | phytochrome interacting factor 3 | ||||



