 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC041431.20 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 501aa MW: 55078.9 Da PI: 6.6169 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.7 | 4.3e-13 | 357 | 403 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI++Lq
EcC041431.20 357 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYINDLQ 403
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 6.8E-42 | 46 | 229 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.028 | 353 | 402 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.02E-18 | 356 | 423 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.77E-15 | 356 | 407 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.1E-18 | 357 | 417 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-10 | 357 | 403 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.4E-15 | 359 | 408 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0004867 | Molecular Function | serine-type endopeptidase inhibitor activity | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 501 aa Download sequence Send to blast |
MGEKFWANED DKRVVESVVG REACELLISS GNAASDVTLP GDLGVQQGLA QLVEGSNWNY 60 AVFWHSASLR SGRITLIWGD GHCCDPKVGA AEDGSSHQNV KSDRVEKKEE LNKLVLQKFH 120 SSFGGSSDDN FAARLNGVSD VEMFYLSSMY FSFQSDSAYG PMGSLKANRV IWASDAPTCL 180 DHYQSRSFLA KSAGFQTVAF VPLKNGVVEL GSVKVIPEEQ KVVEMVRTLL GGPASNKTKV 240 LPKIFGQELS LGGVKPQSVT INFSPKVEDD PMFPSETFDA QRVDSNQFHG GTSNGYPSDN 300 EAKLFSHSNQ IVIGGFSPQT RMSGLEQHKD ESSHHADDRK PRKRGRKPAN GREEPLNHVE 360 AERQRREKLN QRFYALRAVV PNISKMDKAS LLGDAITYIN DLQTKIKVLE TEKDMVNGTH 420 NPQLVPDVQV RHDDTVVTVS CPLEAHPASE VIKTLRNHQV APECDVSMKD GGNEVMHTFS 480 IRTQVGGAEQ LKEKLVAALS K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 1e-29 | 351 | 413 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 338 | 346 | RKPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
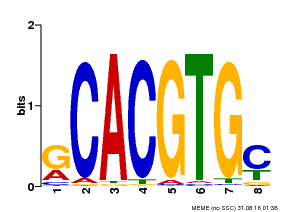
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010038016.1 | 0.0 | PREDICTED: transcription factor bHLH3 | ||||
| Swissprot | O23487 | 1e-170 | BH003_ARATH; Transcription factor bHLH3 | ||||
| TrEMBL | A0A059A6S5 | 0.0 | A0A059A6S5_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010038016.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6611 | 27 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 1e-158 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



