 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC048006.30 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 612aa MW: 67698.7 Da PI: 6.4655 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.5 | 5.1e-13 | 442 | 488 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI++Lq
EcC048006.30 442 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYINELQ 488
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 7.1E-55 | 52 | 240 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF47459 | 6.67E-18 | 437 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.047 | 438 | 487 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.07E-14 | 441 | 492 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-17 | 442 | 500 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.8E-10 | 442 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 444 | 493 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 612 aa Download sequence Send to blast |
MKIELGAGYG PWNDEEKAMV AAVLGTRAFD YLVSRSVSNE NLLMSIGSDE NMQDKLSNLV 60 DHPNSSNFSW NYAIFWQISR SKSGDWLLGW GDGSCREPKD GEESEATRIL NLRLEDEGQQ 120 RMRKRVLDKL HTVFGGSDED NYALGLDRVT DMEMFFLASM YFLFPSGEGG PGKCFASEKH 180 VWLTDALKSS SDYCVRSFLA KSAGIRTIVL VPTDVGVVEL GSVRSVPESS ELVQTIRSSF 240 STNSFMSVKP IAALPMTNEK KDENAPFSNS ALAGKGEAIS KIFGKELTAV NSPGHYREKL 300 AVRKMDSRQS WEPHHNGSKL PFSTPRNGTH DTSWAHAHGV KQLSPVEFYG SQTSASKLEE 360 RMNSGRNDFG LNRYPTPKQV QMQIDFTGAT SRPSVITRPF TADSEHSDVE ASCKEEQGGA 420 ANEKRPRKRG RKPANGREEP LNHVEAERQR REKLNQRFYA LRAVVPNISK MDKASLLGDA 480 IAYINELQAK LKIMEADRER FGSTSRDASD VEVNATAEQS NQSPDVDIQA TAADEVVVRV 540 SCPLNSHPAS RVIQAFQEAQ VTVLDSKLST ANDTVFHTFL IKSQGSEQMT KEKLTAAFSR 600 ESNSLEPLSS VG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 3e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 423 | 431 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
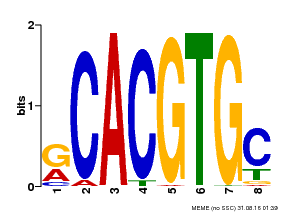
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010055004.1 | 0.0 | PREDICTED: transcription factor bHLH13 | ||||
| Swissprot | A0A3Q7ELQ2 | 0.0 | MTB1_SOLLC; Transcription factor MTB1 | ||||
| TrEMBL | A0A059C0Q7 | 0.0 | A0A059C0Q7_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010055004.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



