 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC053822.30 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 450aa MW: 49095.2 Da PI: 9.4719 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.6 | 2.2e-16 | 58 | 102 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++g ++t+ q++s+ qk+
EcC053822.30 58 RERWTEEEHKKFLEALKLYGRA-WRRIEEYVG-TKTAVQIRSHAQKF 102
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.27E-16 | 52 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.148 | 53 | 107 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 2.5E-17 | 56 | 105 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-13 | 57 | 105 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-9 | 58 | 100 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.7E-14 | 58 | 101 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.69E-10 | 60 | 103 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006011 | Biological Process | UDP-glucose metabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0015992 | Biological Process | proton transport | ||||
| GO:0046034 | Biological Process | ATP metabolic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003983 | Molecular Function | UTP:glucose-1-phosphate uridylyltransferase activity | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0016765 | Molecular Function | transferase activity, transferring alkyl or aryl (other than methyl) groups | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 450 aa Download sequence Send to blast |
MQDQDGSAHS SNMTVGSNEM SLSDSLHSIT SISLKEQFSC ANDYAPKVRK PYTITKQRER 60 WTEEEHKKFL EALKLYGRAW RRIEEYVGTK TAVQIRSHAQ KFFSKVVKDP ADGKVETIEI 120 PPPRPKRKPM HPYPRKLLAH PINKESLPQE QPIRSGSPNF SVSELENQSP MSVLSTLGSD 180 GLGSSGASTP SGSSSPSSSG GVACRAGLSL FESPAEENGP ITSEKLELHA RGCEASGGAK 240 EEAPSRTLKL FGMTVSVTDS HWQSSSAVKS ESKEMKLQDP LSQDTLSATS PAKTNRVLNL 300 VQPRDTSTDH MEAGPAPSAP WLNLYGNLTI SLSPIRNKVA PRSTCGDINK NGIPESRNGV 360 TSNEFHSGFD LKLSFLSKPS STSVAPELMT RQERRGRGFM PYKRCMAEKE TQPSGATGLE 420 REVQRVRLSL ECASKLVESA SPLGPSNRIR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Morning-phased transcription factor integrating the circadian clock and auxin pathways. Binds to the evening element (EE) of promoters. Does not act within the central clock, but regulates free auxin levels in a time-of-day specific manner. Positively regulates the expression of YUC8 during the day, but has no effect during the night. Negative regulator of freezing tolerance. {ECO:0000269|PubMed:19805390, ECO:0000269|PubMed:23240770}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
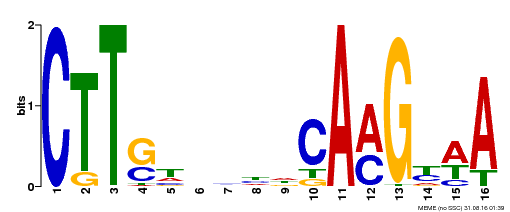
| Motif ID | Method | Source | Motif file |
| MP00212 | DAP | Transfer from AT1G65910 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of transcript abundance near subjective dawn. Down-regulated and strongly decreased amplitude of circadian oscillation upon cold treatment. {ECO:0000269|PubMed:19805390, ECO:0000269|PubMed:23240770}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010062320.1 | 0.0 | PREDICTED: protein REVEILLE 2 | ||||
| Swissprot | F4KGY6 | 1e-50 | RVE1_ARATH; Protein REVEILLE 1 | ||||
| TrEMBL | A0A059BV06 | 0.0 | A0A059BV06_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010062320.1 | 0.0 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 6e-40 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



