 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC053990.70 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 710aa MW: 81960.3 Da PI: 8.0111 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 75.5 | 1.1e-23 | 88 | 173 | 1 | 90 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHela 90
+fY+eYA++vGF +++++s++sk++g++++ + Cs+ g+++e++++ ++r+ +t+Cka +++k+++dg w ++++++eHnH++
EcC053990.70 88 SFYREYARSVGFGITIKASRRSKKSGKFIDVKIACSRFGSKRESSST----VHPRPCMKTNCKAGMNMKRREDGIWIIYSVVKEHNHDID 173
5*****************************************99887....99**********************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 6.7E-21 | 88 | 173 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 2.2E-27 | 256 | 349 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 10.027 | 536 | 572 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 2.1E-6 | 545 | 570 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.4E-5 | 547 | 574 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0055114 | Biological Process | oxidation-reduction process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005506 | Molecular Function | iron ion binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0016705 | Molecular Function | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | ||||
| GO:0020037 | Molecular Function | heme binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 710 aa Download sequence Send to blast |
MPAEEEDVNS HEGSENILVA PHCNSGENVN SHNGGEDMMG APQHDSEEND ACIMNQVGTN 60 SIVDVHHRLL STDPRIGLEF QTKEAAYSFY REYARSVGFG ITIKASRRSK KSGKFIDVKI 120 ACSRFGSKRE SSSTVHPRPC MKTNCKAGMN MKRREDGIWI IYSVVKEHNH DIDPDDFYNS 180 LWGRTKQSGA LAYQKKGLQA FLNEEDTQVM LEHFMSMQQE NPNFFYAVDL GPDKCLRNVF 240 WVDAKGRHDY GSFCDVVYFD IFYVKEKYGI PFLPIVGVNH HAQYLLLGCA LIGAEGKLSY 300 SWVMQAWLNA MGHQAPKVII TDQDKQLKEA VADVFPDIRH CFCLWHIFRK IPLTLAHVLD 360 KYDKFMDKFN KCVYLSWTEE QFEKRWQKLI HKFEVGEDEW VCSLYEDRKK WVPTYMQDAF 420 LAGMSSADRT RSMTAFFDSY LYRGISFKDF LKQYKVYLQE RYEREAASDA ESRYKPPVLR 480 SALTFEKQLS TIYTNAVFKQ FQDEVLGVEL CHLQRENGNE PAAIFRVEEH RKSQNFFVSW 540 KEVDAEILCS CKSFEYRGFL CRHAMLVLQT SGVSSIPCQY ILKRWTKNAK IRETESEKSK 600 ELLASRMQRF NDLCKRAIKL GEQGSLSQEA YNIACQALEE VSRQYSRVNS RWSVSEPGLV 660 AAHQNAEAPE ENNRNVVKSL KRKKICKKRK VSLRLEFDTL SSKCVAATVT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription activator involved in regulating light control of development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
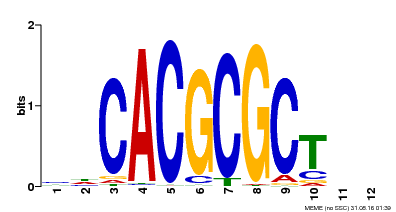
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated in hypocotyls by far-red light treatment. {ECO:0000269|PubMed:15591448}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018727910.1 | 0.0 | PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X2 | ||||
| Swissprot | Q3EBQ3 | 0.0 | FRS2_ARATH; Protein FAR1-RELATED SEQUENCE 2 | ||||
| TrEMBL | A0A059DCQ8 | 0.0 | A0A059DCQ8_EUCGR; Uncharacterized protein (Fragment) | ||||
| STRING | XP_010044813.1 | 0.0 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32250.3 | 0.0 | FAR1-related sequence 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



