 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC053990.90 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 784aa MW: 89921 Da PI: 7.3697 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 85.6 | 7.7e-27 | 89 | 191 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtkleleH 85
+fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ +++ +ra +t+Cka+++vk+++dgkw+++++++eH
EcC053990.90 89 SFYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYEKSfnrprarqskqdPDNATGRRACAKTDCKASMHVKRRSDGKWVIHTFVKEH 185
5******************************************9999999998776655444555999***************************** PP
FAR1 86 nHelap 91
nHel p
EcC053990.90 186 NHELLP 191
***975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.3E-24 | 89 | 191 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.8E-29 | 289 | 380 | IPR018289 | MULE transposase domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0055114 | Biological Process | oxidation-reduction process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005506 | Molecular Function | iron ion binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0016705 | Molecular Function | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | ||||
| GO:0020037 | Molecular Function | heme binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 784 aa Download sequence Send to blast |
MDIDLRLPSG GHEKEEEEPH GIDNMLDGDE KLHNGDMVTT TLADLGDVSH ASDGGALNPP 60 TADMMEVKEY MNLEPLSGME FESHGEAYSF YQEYARSMGF NTAIQNSRRS KTSREFIDAK 120 FACSRYGTKR EYEKSFNRPR ARQSKQDPDN ATGRRACAKT DCKASMHVKR RSDGKWVIHT 180 FVKEHNHELL PAQAISEQTR KMYAAMARQF AEYKNVVGLK NDTRNPFEKG RNILLEGGDA 240 RILLEFFSQM QCSNSNFFHA VDLADDQRIR SLLWVDAKSR HDYSSFSDVI SFDTTYIRSK 300 YKMPLALFVG VNQHYQFMLL GCALLSDESA ATYSWLMQTW LKAVGGQHPK AIITDQDKTI 360 KTVISDVFHS SRHCFCLWQI WGKLSENLSH VIKRHDNFMA KFEKCVNRSW TEAEFEKRWW 420 KIINRCELRE DEWMHSLYED RRHWVPTFLK DAFFAGMCSI QRADSVNAFF DKYVHKKTTX 480 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 540 XXXXXXXXXX XXXXXXAKNR LLMTEESEQV QSRVQRYNDL CQRAMRLSEE GSLSQEAYSL 600 ASRAIEEVFG NCIGQNTISK GLAEAGTSAT HSLLCAEDDS QVRGMSKTNK KKNPTKKRKV 660 TSEPDIMTIG AQESLQQMEK LTPRAVTTLD SYYGTQQSVP GMVQLNLMAP TRDNYYGSQQ 720 TIQGLGQLNS IAPSHDGYYT PQQSMHGLGQ MDFFRSPGGF TYSIRQDDSN VRAAQLHEDA 780 SRHA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
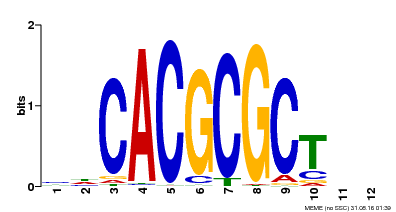
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010044783.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Refseq | XP_010044788.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Refseq | XP_018727883.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A059DDJ6 | 0.0 | A0A059DDJ6_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010044783.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM118 | 26 | 253 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



