 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC054041.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 366aa MW: 39826.5 Da PI: 8.3844 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.2 | 0.00027 | 11 | 33 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y+C C+k+Fs+ L H r+H
EcC054041.10 11 YVCDFCNKRFSSGKGLGGHKRVH 33
9**************99999998 PP
| |||||||
| 2 | zf-C2H2 | 18.9 | 4e-06 | 76 | 97 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C Cgk+F++ +L H+r+H
EcC054041.10 76 TCLMCGKTFPSMKSLFGHMRSH 97
69*******************9 PP
| |||||||
| 3 | zf-C2H2 | 19.4 | 2.9e-06 | 260 | 282 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC++C+ksFs++ L H+++H
EcC054041.10 260 FKCNICNKSFSTHQALGGHMSSH 282
89*******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 1.3E-6 | 8 | 36 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-7 | 8 | 35 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.531 | 11 | 38 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF13912 | 4.9E-8 | 11 | 33 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.021 | 11 | 33 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 13 | 33 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 10.076 | 75 | 102 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0049 | 75 | 97 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 7.48E-10 | 76 | 97 | No hit | No description |
| PROSITE pattern | PS00028 | 0 | 77 | 97 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF13912 | 1.4E-6 | 77 | 99 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.48E-10 | 255 | 282 | No hit | No description |
| Pfam | PF13912 | 7.3E-10 | 259 | 283 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 10.949 | 260 | 283 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.4E-5 | 260 | 284 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.001 | 260 | 282 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 262 | 282 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006821 | Biological Process | chloride transport | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0055085 | Biological Process | transmembrane transport | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005247 | Molecular Function | voltage-gated chloride channel activity | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MQGDVQLENP YVCDFCNKRF SSGKGLGGHK RVHTQASKRN HQFLHKNIHK PKFKKSGGVD 60 DAYLSKAKKS HDIDATCLMC GKTFPSMKSL FGHMRSHPDR EWRGIHPPAR SNHSMSVSDS 120 AGPCRVHDRA TVHPALPQST LGSLRQISSE ALLQSTSGRV TEGKGKGKGK GKGKMDEFQG 180 CDGDADDKGS THERSNEAES NCEDGEYKST QYQLELNILV EKGNIKKKRR KRLPRNLEAG 240 AEVACCINHS RKISKTPERF KCNICNKSFS THQALGGHMS SHTKVTIKKS RSGDDHKGSA 300 SADLEADERA VAAAATQEAE TDGGADAQCL GDSSGDGAPS VQALDCKMLV IDLNELPPMD 360 EDEQEG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 225 | 229 | KKKRR |
| 2 | 225 | 230 | KKKRRK |
| 3 | 225 | 231 | KKKRRKR |
| 4 | 226 | 230 | KKRRK |
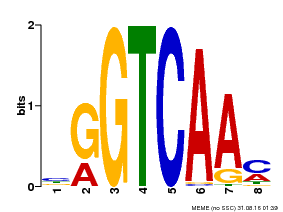
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| STRING | XP_010064659.1 | 1e-104 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56200.1 | 5e-17 | C2H2 family protein | ||||



