 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC054602.20 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1402aa MW: 157118 Da PI: 9.331 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 10.8 | 0.0016 | 1287 | 1312 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
ykC C++sF +k +L+ H ++ +
EcC054602.20 1287 YKCDleGCTMSFGSKQELTLHKKNvC 1312
99*******************99866 PP
| |||||||
| 2 | zf-C2H2 | 13.4 | 0.00023 | 1311 | 1334 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+Cp Cgk F ++ +L++H r+H
EcC054602.20 1311 VCPvkGCGKKFFSHKYLVQHRRVH 1334
69999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 10.9 | 0.0014 | 1370 | 1396 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y C Cg++F+ s++ rH r+ H
EcC054602.20 1370 YICAepGCGQTFRFVSDFSRHKRKtgH 1396
789999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.8E-16 | 20 | 61 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.34 | 21 | 62 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.5E-14 | 22 | 55 | IPR003349 | JmjN domain |
| SMART | SM00558 | 4.3E-46 | 193 | 362 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.213 | 196 | 362 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.57E-25 | 207 | 377 | No hit | No description |
| Pfam | PF02373 | 1.3E-36 | 226 | 345 | IPR003347 | JmjC domain |
| SMART | SM00355 | 11 | 1287 | 1309 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.466 | 1310 | 1339 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-5 | 1310 | 1338 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0037 | 1310 | 1334 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1312 | 1334 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.94E-9 | 1326 | 1368 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.1E-8 | 1339 | 1364 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1340 | 1364 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.741 | 1340 | 1369 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1342 | 1364 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.32E-8 | 1358 | 1392 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.4E-9 | 1365 | 1393 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.79 | 1370 | 1396 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.889 | 1370 | 1401 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1372 | 1396 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006165 | Biological Process | nucleoside diphosphate phosphorylation | ||||
| GO:0006183 | Biological Process | GTP biosynthetic process | ||||
| GO:0006228 | Biological Process | UTP biosynthetic process | ||||
| GO:0006241 | Biological Process | CTP biosynthetic process | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0004550 | Molecular Function | nucleoside diphosphate kinase activity | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0016747 | Molecular Function | transferase activity, transferring acyl groups other than amino-acyl groups | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1402 aa Download sequence Send to blast |
MAAATEPSQE VFSWLKNLPL APEYHPTLAE FQDPIAYIFK IEKEASKYGI CKIVPPVPPG 60 PKKSAITNLN RSLAARASSS ERPNSKSSPT FTTRQQQIGF CPRKPCPVKK PVWQSGENYT 120 FQDFETKAKS FERVYLRKFY KKGSPSALEI ETLYWKAAVD KPFSVEYAND IPGSAFPPIR 180 SKKGRETGEG MLTVGETNWN MRGVSRSKGS LLRFMKEEIP GVTSPMVYVA MMFSWFAWHV 240 EDHDLHSLNY LHMGAGKTWY GVPRDAAVAF EEVVRVQGYG GEVNPLITFA VLGEKTTVMS 300 PEVLVGAGVP CCRLVQNAGE FVVTFPRAYH SGFSHGFNCG EAANIATPEW LLFAKEAAIR 360 RASINYPPMV SHFQLLYDLA LAFCSRVPTC INTGPRSSRL KDKKKGEGES MVKEQFVQNC 420 IENNKLLHIL GKDSTVVRLP ESSSDISVCS NLRVGSHLRV NRSLSEDTMT SHGGLVSDNL 480 IADRKQTIDL VDDLYSMKSF SSFYERKRIL SMYGAGDIQP PRDGDGDSKA QGDRLSDQRL 540 FSCVTCGILT FACAAIVQPR EPAARYLMSA DCSFFNDWTV NPGVPTKGFN VSAGADIIAY 600 KRRTQAGSVE KSRAAALYDV SPLQSTNYQI QEAHQSSEAF SNSDKLKGTS ALGLLALNYG 660 NSSDSEEDQI DSDAPNETTS RPQYGNNNLP DSALPPFLPE HHSGPNGSSP HNCCPEIGYR 720 GANFVDKSHQ TFKYSANFRA DETRSSEKHG STGFPKSDED SSRMHIFCLE HAIEVDQRLR 780 PIGGVYIYLV CHPDYPRVEA EAKLLAEELG IDHSWNEIAF RDAMGEDKER IQSALDSEEA 840 IAGYGDWAVK LGINLFYSAN LCSSPLYSKQ MPYNSVIYNA FGRESSSSPA KLDDFGRRPS 900 KQKKSVAGKW CGKVWMSNQV HPFLAPKDAE EVEEERSFHA SMTSDDKLER QSGLNRETTL 960 ATRKFSRKRK MARESGPSKK KSVSRKEEVS DDALAEDSEK LMSIPKHKTA KSFRREDPVS 1020 DDQVDEISCL QHQTIPRRKQ KKFVKRGYAS SDDDSLEDKF KALRGKNFRK ATNFTSSGDM 1080 LSDDSLEEDS QQQGRILTGK KTKHYERDDV MSDDSEGNYN KLQHPRIRKK TPANAKFEGA 1140 DSDSSPEDAF PQRQRSLRNR KVNRDTLRIK NQGNLLEKLK KRGIPRVTKE VGARHLKQES 1200 SRSRSNKYER SVEQSDESDE EDPEGGGPST RLRKRTAKPT KERKADKPPP GKKQANNGKK 1260 NVKAGPTVKR PVGRNDARME KFEEAEYKCD LEGCTMSFGS KQELTLHKKN VCPVKGCGKK 1320 FFSHKYLVQH RRVHLDDRPL KCPWKGCKMT FKWAWARTEH IRVHTGARPY ICAEPGCGQT 1380 FRFVSDFSRH KRKTGHSVKG KG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 1e-78 | 14 | 458 | 8 | 421 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 1e-78 | 14 | 458 | 8 | 421 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 965 | 970 | SRKRKM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
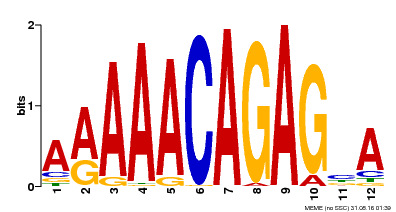
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010037473.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A059A5S0 | 0.0 | A0A059A5S0_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010037473.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



