 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcC079473.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 184aa MW: 19118.2 Da PI: 7.2658 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.1 | 1.2e-16 | 124 | 170 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L++l+P++ K +Ka++L +A+eY+k+Lq
EcC079473.10 124 VHNLSEKRRRSRINEKMKALQSLIPNS-----NKTDKASMLDEAIEYLKQLQ 170
5*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 6.2E-19 | 117 | 175 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.57E-19 | 117 | 176 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.841 | 120 | 169 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.82E-14 | 123 | 174 | No hit | No description |
| Pfam | PF00010 | 3.3E-14 | 124 | 170 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.3E-18 | 126 | 175 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 184 aa Download sequence Send to blast |
PDEISLFLRQ VLQRSGRRAE NLPGRPGGGS GGGGGGGGGG ISFDRVVDSF SASASASGGG 60 PLAGNARLSA VGAAAAASSS VGASENEADE YDCESEGLEA LLEDVPARPN PPRGSSKRSR 120 TAEVHNLSEK RRRSRINEKM KALQSLIPNS NKTDKASMLD EAIEYLKQLQ LQVQVIALSQ 180 ILQC |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that plays a role in floral organogenesis. Promotes the growth of carpel margins and of pollen tract tissues derived from them. {ECO:0000269|PubMed:10225997, ECO:0000269|Ref.8}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
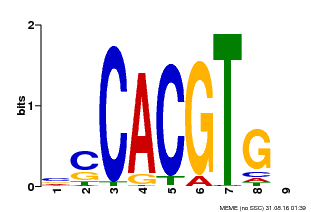
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by the A class gene AP2 in the first whorl and by ARF3/ETT in gynoecium. {ECO:0000269|PubMed:11245574}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010030576.1 | 6e-82 | PREDICTED: transcription factor SPATULA | ||||
| Swissprot | Q9FUA4 | 1e-33 | SPT_ARATH; Transcription factor SPATULA | ||||
| TrEMBL | A0A2I4GM63 | 8e-56 | A0A2I4GM63_JUGRE; transcription factor SPATULA-like isoform X2 | ||||
| TrEMBL | A0A2I4GM66 | 4e-55 | A0A2I4GM66_JUGRE; transcription factor SPATULA-like isoform X1 | ||||
| STRING | XP_010030576.1 | 2e-81 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4199 | 27 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 4e-35 | bHLH family protein | ||||



