 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcS566401.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 307aa MW: 34267.4 Da PI: 6.2637 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 158.3 | 5.4e-49 | 27 | 164 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq.ss 97
g+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve+DGttyr++ p + +++ +s s+ + s
EcS566401.10 27 GKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWTVEADGTTYRQSPPPS----QLATFSARSMDSPASaAS 119
689******************************************************************776555....666777777777777789 PP
DUF822 98 lkssalaspvesy.saspksssfpspssldsislasa.......a 134
lks+ +++ ++s+ sa+++ ++++sp+slds+ +a++
EcS566401.10 120 LKSYGVKTTLDSQpSAVMRMDESLSPASLDSVVIAEKdpksvkyG 164
*******999988467899***************98755444441 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 7.0E-50 | 27 | 170 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 4.7E-36 | 197 | 307 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 4.96E-31 | 201 | 307 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 1.1E-17 | 218 | 305 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-13 | 233 | 247 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-13 | 254 | 272 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-13 | 276 | 297 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
QQQQQQQQRR PRGFAATAAA AATAGPGKGK REREKEKERT KLRERHRRAI TSRMLAGLRQ 60 YGNFPLPARA DMNDVLAALA REAGWTVEAD GTTYRQSPPP SQLATFSARS MDSPASAASL 120 KSYGVKTTLD SQPSAVMRMD ESLSPASLDS VVIAEKDPKS VKYGNASPIN SVIECLDAEQ 180 LIHDVSSVDH QNVFTDTEFV PVYVKLDGTI NNYCQLVDQE VVRQELNHIK SLNADGVVVD 240 CWWGIVEGWN PQRYVWSGYR ELFHIIREFK LKLQVVLAFH EYGGNDSGDF SVSLPQWVLE 300 IGKENED |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1fa2_A | 2e-24 | 199 | 307 | 12 | 121 | BETA-AMYLASE |
| 5wqs_A | 2e-24 | 199 | 307 | 12 | 121 | Beta-amylase |
| 5wqu_A | 1e-24 | 199 | 307 | 12 | 121 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
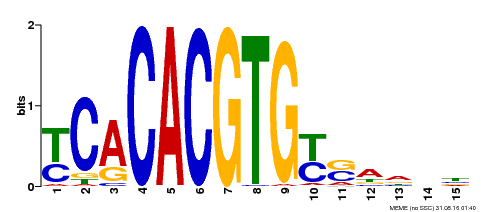
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010054915.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 1e-123 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A059CJR3 | 0.0 | A0A059CJR3_EUCGR; Beta-amylase | ||||
| STRING | XP_010054915.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 1e-117 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



