 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcS751352.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 184aa MW: 21087.6 Da PI: 5.51 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 58 | 111 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++++q+++Le+ Fe ++++ e++++LA++l+L+ rqV +WFqNrRa++k
EcS751352.10 58 KKRRLSANQVKALEKNFEIENKLEPERKARLAEELSLQPRQVAIWFQNRRARWK 111
56689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.4 | 1.4e-41 | 57 | 149 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrls++qvk+LE++Fe e+kLeperK++la+eL lqprqva+WFqnrRAR+ktkqlE+dy +Lk++yd+lk + ++Le+e+e+L++el+e
EcS751352.10 57 EKKRRLSANQVKALEKNFEIENKLEPERKARLAEELSLQPRQVAIWFQNRRARWKTKQLERDYGFLKASYDSLKLDFDSLEQEKESLAAELTE 149
69**************************************************************************************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.13E-19 | 51 | 115 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.038 | 53 | 113 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.6E-16 | 56 | 117 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 6.7E-16 | 58 | 111 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.35E-14 | 58 | 114 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-19 | 60 | 120 | IPR009057 | Homeodomain-like |
| PROSITE pattern | PS00027 | 0 | 88 | 111 | IPR017970 | Homeobox, conserved site |
| Pfam | PF02183 | 8.8E-18 | 113 | 154 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 184 aa Download sequence Send to blast |
MKRLSSSDSS DVWISMCSGK EEKVLKKSQG YSIEFQAMLD SLDQEDHSGE EAGLITEKKR 60 RLSANQVKAL EKNFEIENKL EPERKARLAE ELSLQPRQVA IWFQNRRARW KTKQLERDYG 120 FLKASYDSLK LDFDSLEQEK ESLAAELTEL KVKLRRETSE SSNHSAVKQE SPLSESSEDG 180 KPGS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 82 | 88 | ERKARLA |
| 2 | 105 | 113 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may function as a negative regulator of the flowering time response to photoperiod. May act to repress cell expansion during plant development. {ECO:0000269|PubMed:14623244}. | |||||
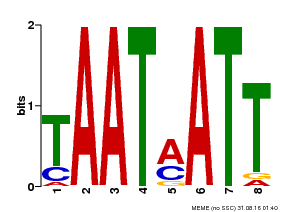
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00051 | PBM | Transfer from AT4G40060 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010060054.1 | 1e-124 | PREDICTED: homeobox-leucine zipper protein ATHB-6 isoform X3 | ||||
| Swissprot | Q940J1 | 3e-53 | ATB16_ARATH; Homeobox-leucine zipper protein ATHB-16 | ||||
| TrEMBL | A0A059BKG0 | 1e-123 | A0A059BKG0_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010060054.1 | 1e-124 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM545 | 28 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G40060.1 | 3e-52 | homeobox protein 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



