 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | EcS783656.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 132aa MW: 15139.5 Da PI: 8.738 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 66.9 | 3e-21 | 29 | 119 | 4 | 98 |
E-..-HHHHTT-EE--HHH.HTT.---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 4 vltpsdvlksgrlvlpkkfaeeh.ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
vl ps+ + ++++lp fae+h +g++ ++l++++g++W v++ yr+++ l++GW +F +n+L+egD++vF+l++ +e l v+vfr
EcS783656.10 29 VLRPSYLYRGCIMYLPSCFAEKHlNGVSG---FIKLQSTDGKQWPVRCLYRGGR--AKLSQGWYDFSLENNLEEGDVCVFELMRAREVVLQVTVFR 119
7889*******************877744...5***************665555..66*************************9988889999998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 3.3E-26 | 17 | 119 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 6.8E-27 | 19 | 119 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.85E-25 | 24 | 119 | No hit | No description |
| PROSITE profile | PS50863 | 14.24 | 26 | 121 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.9E-20 | 26 | 121 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 9.6E-19 | 29 | 119 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0010048 | Biological Process | vernalization response | ||||
| GO:0034613 | Biological Process | cellular protein localization | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 132 aa Download sequence Send to blast |
KRLVTAEERE KAINAAKMFE PPNPFCRVVL RPSYLYRGCI MYLPSCFAEK HLNGVSGFIK 60 LQSTDGKQWP VRCLYRGGRA KLSQGWYDFS LENNLEEGDV CVFELMRARE VVLQVTVFRV 120 VEEGGLLSHH GR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4i1k_A | 2e-66 | 1 | 122 | 24 | 144 | B3 domain-containing transcription factor VRN1 |
| 4i1k_B | 2e-66 | 1 | 122 | 24 | 144 | B3 domain-containing transcription factor VRN1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the regulation of vernalization. Acts as transcriptional repressor of FLC, a major target of the vernalization pathway. Binds DNA in vitro in a non-sequence-specific manner. {ECO:0000269|PubMed:12114624}. | |||||
| UniProt | Involved in the regulation of vernalization (By similarity). Binds to DNA in vitro in a non-sequence-specific manner. XLG2 promotes the DNA binding activity of RTV1 specifically to promoter regions of FT and SOC1 in vivo and thus leads to the activation of floral integrator genes. {ECO:0000250, ECO:0000269|PubMed:22232549}. | |||||
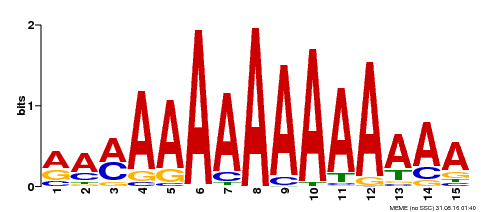
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00192 | DAP | Transfer from AT1G49480 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold. {ECO:0000269|PubMed:22232549}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010066068.1 | 1e-93 | PREDICTED: B3 domain-containing transcription factor VRN1 | ||||
| Swissprot | Q8L3W1 | 5e-64 | VRN1_ARATH; B3 domain-containing transcription factor VRN1 | ||||
| Swissprot | Q9XIB5 | 6e-65 | REM19_ARATH; B3 domain-containing protein REM19 | ||||
| TrEMBL | A0A088AWV0 | 3e-77 | A0A088AWV0_SOYBN; Reduced vernalization response 1 | ||||
| STRING | XP_010066068.1 | 4e-93 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1124 | 28 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49480.1 | 2e-67 | related to vernalization1 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



