 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.A00102.1.p | ||||||||
| Common Name | EUGRSUZ_A00102, LOC104435578 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 420aa MW: 45742.7 Da PI: 6.4604 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.3 | 2.2e-10 | 224 | 282 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i+eLe+kv++L++e ++L +l l+ l++e+
Eucgr.A00102.1.p 224 KRAKRIWANRQSAARSKERKMRYIAELERKVQTLQTEATTLSAQLTLLQRDTNGLTAEN 282
9*********************************************9998888777776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.2E-15 | 220 | 284 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.898 | 222 | 285 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 9.85E-11 | 224 | 275 | No hit | No description |
| Pfam | PF00170 | 5.9E-9 | 224 | 278 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.8E-10 | 224 | 278 | No hit | No description |
| CDD | cd14703 | 1.62E-25 | 225 | 274 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 420 aa Download sequence Send to blast |
MDKDKLTGHS SGLPPPPSGR FAGFSPNANA FSVKPEQPSA AASSSSFLPM AQGQNLVSDF 60 SHDISRMPDN PPRNRGHRRA HSEILTLPDD ISFDSDLGVV GAADGPSFSD DTEEDLFSMY 120 LDMDKFNSSS ATSSFQMGEP SSAPMAMPGA GDSGSGAAPS SAENVASGSN ERPRIRHQHS 180 QSMDGSTSIK PEMLMSGSED ASAADAKKAM SAAKLAELAL IDPKRAKRIW ANRQSAARSK 240 ERKMRYIAEL ERKVQTLQTE ATTLSAQLTL LQRDTNGLTA ENSELKLRLQ TMEQQVHLQD 300 ALNEALKEEI QHLKVLTGQS MPNGGPMMNY PSFGSGQQFY PNNHAMHTLL AAQQFQQLQI 360 QSQKQQHQFQ QHQLHHQLQQ QQLQHEQQQP AGDLKFRAPI PSPNQKDGNA AESNSSKME* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
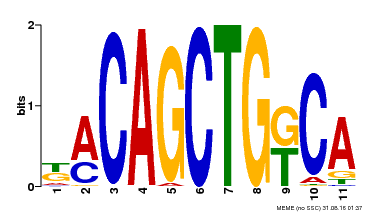
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.A00102.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010046685.1 | 0.0 | PREDICTED: probable transcription factor PosF21 isoform X2 | ||||
| Swissprot | Q04088 | 1e-159 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A059DB53 | 0.0 | A0A059DB53_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010046629.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2445 | 27 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-136 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.A00102.1.p |
| Entrez Gene | 104435578 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



