 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.A01019.1.p | ||||||||
| Common Name | EUGRSUZ_A01019, LOC104434497 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 984aa MW: 109055 Da PI: 6.1144 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 133.9 | 5.4e-42 | 153 | 230 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adls+ak+yhrrhkvCe+hska+++lv +++qrfCqqCsrfh+l+efDe+krsCrrrLa+hn+rrrk+++
Eucgr.A01019.1.p 153 VCQVEDCGADLSNAKDYHRRHKVCEMHSKATKALVGNIMQRFCQQCSRFHALQEFDEGKRSCRRRLAGHNKRRRKTNP 230
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.4E-34 | 147 | 215 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.334 | 151 | 228 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.02E-38 | 152 | 232 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.8E-30 | 154 | 227 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 1.03E-6 | 754 | 869 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.8E-5 | 757 | 872 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.05E-5 | 758 | 868 | No hit | No description |
| PROSITE profile | PS50088 | 8.816 | 809 | 832 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 8.72 | 809 | 832 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 984 aa Download sequence Send to blast |
MEARVEAQAP RHFYGLPPVD LRALGKRSVE WDSNDWKWDG DLFVARPLNP VPSDFPGRQF 60 LPLENGIPAA AGNSSNSSSS CSDEVNLRTD DGKRELEKRR RVIVVEDDNS DDKAGGLTLK 120 LGGYPVADRD VGTWEGNSGK KTKLAGGSAS RAVCQVEDCG ADLSNAKDYH RRHKVCEMHS 180 KATKALVGNI MQRFCQQCSR FHALQEFDEG KRSCRRRLAG HNKRRRKTNP EATPNANSVN 240 DEQASSYLLI SLLKILTSIH SNQSKQATDQ DLLAHILRSI ASSTSGHGER NISGLLQESQ 300 AILNGGTSLA NSELVHPMSS KGVEGPSRLV QPQAAGPASQ ILANGREATG GQSKLNNFDL 360 NDTYIDSDDG MEDLDRSPVP ANLGTSSIDC PSWMQQESHQ SSPPQTSGTS DSASAQSPSS 420 SSGDTQSRTD RIVFKLFGKD PNDFPLLLRT QILDWLSHSP TDIESYIRPG CIVLTVYLRQ 480 DEDAWEELCY DLSSSLNRLL DVSDDFFWKT GWMYMRMQHQ MAFIYNGQVV VDTSLCLRSM 540 NCSRIMSIRP IATSSSETTQ FSVKGFNMCH PATRLLCALE GTYLVQEATL ELMNEEDSLE 600 EHDELQCVKF SCSIPRVTGR GFIEVENGGL PGSFFPFIVA ESDVCLEIRS LENALEFVEA 660 DADDHGTSKF AAKNQAFEFI QEMGWILHRS GLRSRLRQLD PYTDSFSFKR FKWLIEFSMD 720 HDWCAVVKKL LDILFDGTVG AGEHPSIYHA VLEMGLVHRA VRRNCRPLVQ LLLRYIPQSV 780 SEKLESELKS LISGSHQNFL FRPDAVGPAG LTPLHIAAGK DDSEDVLDAL IDDPGMVGIE 840 AWKTARDSTG STPEDYARLR GHYSYIHLVQ KKINKRPSVP HVVLDIPDAV PDSNGVQKQN 900 KETTSSFEIG KTEQQLAHKS CKLCSQKVAY GMRSRSLLYR PTMLSMVGIA AVCVCVALLF 960 KSCPEVLYVF QPFRWEALDF GSS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-31 | 145 | 227 | 2 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 97 | 101 | KRRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
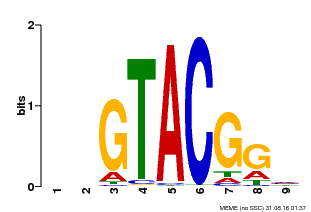
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.A01019.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010045710.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | A0A059DE14 | 0.0 | A0A059DE14_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010045710.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.A01019.1.p |
| Entrez Gene | 104434497 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



