 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.A02043.1.p | ||||||||
| Common Name | EUGRSUZ_A02043 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 569aa MW: 62335.1 Da PI: 7.8324 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.4 | 9.3e-17 | 376 | 422 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+ksLq
Eucgr.A02043.1.p 376 VHNLSERRRRDRINKKMKALQELIPHC-----NKTDKASMLDEAIEYLKSLQ 422
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.22E-20 | 370 | 434 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.222 | 372 | 421 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.41E-17 | 375 | 426 | No hit | No description |
| Pfam | PF00010 | 3.1E-14 | 376 | 422 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.2E-20 | 376 | 430 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.6E-18 | 378 | 427 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 569 aa Download sequence Send to blast |
MRPGSENLKI RLFHLITLLG TSQRLALKLF VRCAAMNPCL PDWNFEYDLP LTNQKKNIGS 60 DQELVELLWQ NGQVVMHSQN HRKPSSNQHE SSQLPKQDQP IMKGSGSYGN SSNLIQDDET 120 VSWIHYPVED SFEREFCSNF FDELPVPHPM EIDKAVRQVE AEKIVKSASF NDAHNTSNQQ 180 STVKRALVPE ISGNQMPPPR AQFAKSAQQN NVTGNLGKVV NFSQFASPVK GDPRPSSLHF 240 GGDARECSVM TVGSSHCGSN QIVNDPDFSR ASSNAAGTAG FSAGTFKEDV HKNVVSQSEG 300 RKTETLEPTV TSSSGGSDSS FGGTLKESAG ASSHKRKGRD NEESECQSEA AEPEVAGGKK 360 PATRSGSTRR SRAAEVHNLS ERRRRDRINK KMKALQELIP HCNKTDKASM LDEAIEYLKS 420 LQLQLQVMWM GGGMAPMMFP GVQHYMSSMG MTMCPPPMPT VSNNLHLPQV PVVDQSIPMA 480 PNPNQSAICH TPVLNPMNFQ NQMQSPSFPE QFARYMGMHH MQATSQPMNM FKFGPQTVQQ 540 IQTNPLPGSI SGQYSGGATN DALSGKMG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 380 | 385 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
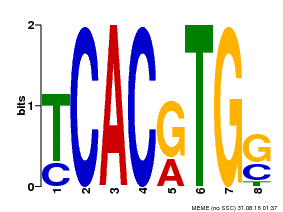
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.A02043.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010054152.1 | 0.0 | PREDICTED: transcription factor PIF5 | ||||
| TrEMBL | A0A059DGA5 | 0.0 | A0A059DGA5_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010054152.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3468 | 25 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 3e-32 | phytochrome interacting factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.A02043.1.p |



