 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.B00645.1.p | ||||||||
| Common Name | EUGRSUZ_B00645 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 598aa MW: 65052.4 Da PI: 8.2239 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.2 | 6.1e-13 | 269 | 307 | 2 | 44 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiL 44
r +h++ E+rRR++iN++f++Lr+l+P++ ++K +Ka+ L
Eucgr.B00645.1.p 269 RSKHSATEQRRRSKINDRFQTLRSLIPNS----DQKRDKASFL 307
889*************************9....8999**9988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 4.2E-19 | 264 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.84E-12 | 266 | 310 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 13.446 | 267 | 317 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.4E-10 | 269 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.44E-10 | 269 | 307 | No hit | No description |
| SMART | SM00353 | 0.0011 | 273 | 355 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.2E-19 | 340 | 368 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 598 aa Download sequence Send to blast |
MMELPQPRPF GTEARKATHD FLSLCSHSTL HHQDPRQPPS QDSFLKTHDF LGKTAAKEEA 60 TSEISSSLER PPPPAPPGPP PSVPPPQSVE HVLPGGIGTY SISHISYFNN QNPPKPEATI 120 YTVAQASSTD RTDENSNCSS YTNSGFTLWE ESAAKKGKTG KENVPSASVR EAEVKLGPWP 180 ASERPSHSSS NNNHRNSFSS LSSSQPSGAK NKSFMDMIRS AKSGSNQEDE LDDDDGEFLL 240 KKETSPIPKG ELRVKVDGKS SDQKANTPRS KHSATEQRRR SKINDRFQTL RSLIPNSDQK 300 RDKASFLLEV LGHSFCSFFP SFLPDRRNLI ILIDEGTAFI QVIEYIQYLQ EKVTNYEGSF 360 QGWSQEPANM MPLRNGQRPT DNYVDQSRNI TSGSSPALVF AAKVDENNIA MAPAVFRKAK 420 NPLEPEMGSV TAFKSVEHHP GMTNRAVPFP MSLQPNISNC VRGSGAVARG DPNLENNASQ 480 PLTQISRIKS LTNECSASTD KLKEEELTIE GGTISISNVY SQGLLNRLTQ ALQSSGVDLS 540 QASISVQIDL GKQANGRNAA SSSLPKDTEA RNQGVTRPRA AGEIADPALK KLKAGKS* |
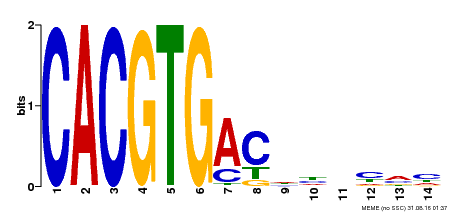
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.B00645.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010031175.1 | 0.0 | PREDICTED: transcription factor BIM1 isoform X1 | ||||
| TrEMBL | A0A059D0L8 | 0.0 | A0A059D0L8_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010031175.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6967 | 27 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 2e-99 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.B00645.1.p |



