 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.B01825.1.p | ||||||||
| Common Name | EUGRSUZ_B01825, LOC104431553 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 661aa MW: 70035.5 Da PI: 5.285 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.1 | 4.9e-16 | 429 | 475 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn Er+RRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Eucgr.B01825.1.p 429 VHNLSERKRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 475
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.64E-17 | 421 | 479 | No hit | No description |
| SuperFamily | SSF47459 | 6.94E-21 | 422 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 7.6E-21 | 422 | 485 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.216 | 425 | 474 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.6E-13 | 429 | 475 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.0E-17 | 431 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 661 aa Download sequence Send to blast |
MPLFELFRRA QEKLNSRQDK GQSCSADLSF IPEDGFVELI WENGQVVMQG QSSRPKKSQP 60 GFGLLSEFTS QSPWISDKDV GNGAASNSKK SEAEPDSGLS EFPMSSPPRE MCLTHDEDIV 120 PWFSCPINEP LQNDYCSDFL PELSGVPVNE LSNLYSFDST QKSNNKEVGP AGVNGAKVPS 180 AADVEINGMG MNTALPSLQQ GVMDAMGSSK CGITYDSTFG DFSRLPRLAV GSTSEKGQAK 240 DATPSAKNSN FINFSHFARV ATPSHANVEN NARASGSGSL ISDRAARKEK CSSTNDSIMA 300 DNLASNLSVC PRTEASSNCQ VQREQLMVAS KPLICQGDAL KNDKSADQSC CASSSKGSPD 360 NEKMTELAAS TSSASTSDAP SCSLKRKCPS TNNSEALAED VDEDSVGVKK AAAAAARSTS 420 SKRSRAAEVH NLSERKRRDR INEKMRALQE LIPNCNKVDK ASMLDEAIEY LKTLQLQVQM 480 MSMANGLYMP PMMFPTAIQR IHPSQMAQFA PMGIGMGLGM GMVGVNCGTH VPVPHVSAST 540 ALHSTAAPNF PLMGLPCQGL PMSMPCLSSL VPFPGVPPLK PGTGSNTSAF VGPGTMENTD 600 AAIPSSVKES TQNVNSASCI TNHTSTQFQA RNGDFQKPTS VQAACPDSDI NVGGVADSVD 660 * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 433 | 438 | ERKRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
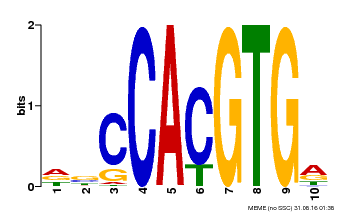
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.B01825.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018723934.1 | 0.0 | PREDICTED: transcription factor PIF3 | ||||
| TrEMBL | A0A059D361 | 0.0 | A0A059D361_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010042473.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7665 | 27 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-51 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.B01825.1.p |
| Entrez Gene | 104431553 |



