 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.B03669.1.p | ||||||||
| Common Name | EUGRSUZ_B03669, LOC104433824 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 432aa MW: 47263.3 Da PI: 7.4135 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.1 | 1.3e-12 | 365 | 410 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR +i +++ +L++l+P+ +k + a++L +Av+YIk+Lq
Eucgr.B03669.1.p 365 SIAERVRRTKISERMRKLQDLVPNM----DKQTNTADMLDLAVDYIKDLQ 410
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.288 | 359 | 409 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.27E-16 | 362 | 424 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.15E-12 | 363 | 414 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.2E-15 | 364 | 420 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-9 | 365 | 410 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-13 | 365 | 415 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 432 aa Download sequence Send to blast |
MESDLQQQQQ QQHHHHAFLD HGHDPPLPQH HQQQQMSSGL SRYRSAPSSF ASLFERDFCN 60 ELLGRPPSPE TERFLARLIS GGQDESPAAA AAAAQNLGGF QRSPPQNEVT NPQPQLMEAV 120 KSEANVIPQQ QSNYSSGSYY QSSSQQPQPP RPNPSSTYGA AGSDRLPPSP KVGGSNPNLV 180 RHSSSPAGLF ANINIDNGFA SMESMRNFVA CGGANAEASF ANANRLTNHM SYSSRSAVGH 240 MSPISELGDK RMEASIDEGA AFGEGNANDY VAGYPMSSWE DSASMSENFN ALKRSRDDDK 300 KFSGLNASDL VNEEVRSRPP MGLTHHLSLP KTSAEISSIE KFLQLQDSVP CKIRAKRGCA 360 THPRSIAERV RRTKISERMR KLQDLVPNMD KQTNTADMLD LAVDYIKDLQ TQVKGLSANR 420 ARCTCLNRQQ Q* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
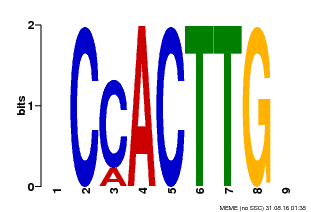
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.B03669.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010045000.1 | 0.0 | PREDICTED: transcription factor bHLH130 isoform X1 | ||||
| TrEMBL | A0A059D976 | 0.0 | A0A059D976_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010045000.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10547 | 26 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 5e-71 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.B03669.1.p |
| Entrez Gene | 104433824 |



