 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.B03926.1.p | ||||||||
| Common Name | EUGRSUZ_B03926 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 420aa MW: 45975.4 Da PI: 9.163 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 23 | 2e-07 | 243 | 265 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C++Cgk F+r nL+ H+r H
Eucgr.B03926.1.p 243 HYCQICGKGFKRDANLRMHMRAH 265
68*******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 5.46E-6 | 241 | 268 | No hit | No description |
| PROSITE profile | PS50157 | 12.632 | 243 | 270 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-6 | 243 | 269 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0028 | 243 | 265 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 245 | 265 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 77 | 304 | 337 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 22 | 342 | 364 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 420 aa Download sequence Send to blast |
MYGVTGHEDI GVVTSSSMDN NAASSTSPAD THSNTLLYNL SLLKDKVHQV QSLISVLVSP 60 PQIAGQSTAT TSMAIASMVT SIQELIVAAS SMMYACQQMS NASASSADNP TSRQELIPRQ 120 QEQQQQQQQQ QQHRIKPVGH GGLLSHHHSQ PTGIYASDQT FDWFADNSYV SSPAMGNNNS 180 GNSNRNNSSR PGQNGRDQAL PQPCGGQRGH NEGSTQGTSL KSGSSNFDII ELDASELLAK 240 YTHYCQICGK GFKRDANLRM HMRAHGDEYK TSSALSNPLK NSSGSNNSIG ADEKECPMKT 300 PRKYSCPQEG CRWNQKHAKF QPLKSMICVK NHYKRSHCPK MYVCKRCNRK QFSVLSDLRT 360 HEKHCGDLKW QCSCGTTFSR KDKLMGHVTL FVGHSPMINL TKPGKVDGQA ASHNVKADR* |
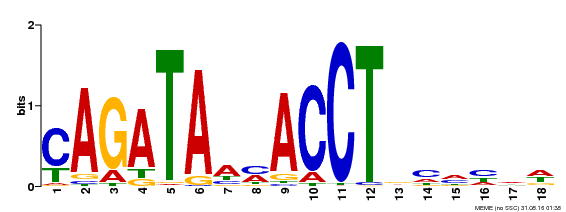
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.B03926.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010045298.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| Refseq | XP_010045299.1 | 0.0 | PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 | ||||
| TrEMBL | A0A059DA80 | 0.0 | A0A059DA80_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010045298.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5752 | 25 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 1e-107 | C2H2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.B03926.1.p |



