 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.D01054.1.p | ||||||||
| Common Name | EUGRSUZ_D01054, LOC104441262 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 324aa MW: 35857.5 Da PI: 9.8896 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 395.4 | 8e-121 | 1 | 323 | 1 | 301 |
GAGA_bind 1 mdddgsre..rnkg.yyepa..aslkenl..glqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalll 86
mdd g+re r+k+ +y++a + l +++ +q+m ++aerda+++ernlalsekkaa+aerd+af+qrd+a+aern+a +erdn++++l++
Eucgr.D01054.1.p 1 MDDGGHREngRHKAdHYKNAqsQWLMQHQpsMKQIMGIMAERDAALQERNLALSEKKAAIAERDIAFHQRDMAIAERNNAYLERDNAIATLQY 93
9*****999*****99995544344444321469*********************************************************** PP
GAGA_bind 87 vensla....salpvgvqvlsgtksidslqq.lse.pqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksek 173
+ensl+ s++p+g+q+++g+k++++ qq +++ + +++ a+++r+++ ++alpi+ a+ea++++++k++++ak ++k+ +k
Eucgr.D01054.1.p 94 RENSLTsgniSSCPPGCQISRGVKHMHHPQQqVHHvSSMSEAAYGTRDMHPSDALPISPVASEATRSRQTKRTKEAKAMSTSKK------APK 180
****9999999******************995556899*******************998888888777775554443322222......222 PP
GAGA_bind 174 skkkvkkesader...............skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrr 251
+ kkvk+e++d + k+++k +dl+ln+v++D+st+P PvCsCtG+ rqCYkWGnGGWqS+CCtttiS+yPLP+++++r
Eucgr.D01054.1.p 181 TPKKVKRETDDLNkllngddgynkpfgvPKSDWKGQDLGLNQVAFDDSTMPSPVCSCTGIARQCYKWGNGGWQSSCCTTTISMYPLPAVPNKR 273
22222222222111333445667899989**************************************************************** PP
GAGA_bind 252 gaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
+aR++grKmS++af klL++LaaeGydls pvDLkdhWAkHGtn+++ti+
Eucgr.D01054.1.p 274 HARVGGRKMSGSAFGKLLSRLAAEGYDLSSPVDLKDHWAKHGTNRYITIK 323
*************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 7.1E-104 | 1 | 323 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 1.7E-172 | 1 | 323 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
MDDGGHRENG RHKADHYKNA QSQWLMQHQP SMKQIMGIMA ERDAALQERN LALSEKKAAI 60 AERDIAFHQR DMAIAERNNA YLERDNAIAT LQYRENSLTS GNISSCPPGC QISRGVKHMH 120 HPQQQVHHVS SMSEAAYGTR DMHPSDALPI SPVASEATRS RQTKRTKEAK AMSTSKKAPK 180 TPKKVKRETD DLNKLLNGDD GYNKPFGVPK SDWKGQDLGL NQVAFDDSTM PSPVCSCTGI 240 ARQCYKWGNG GWQSSCCTTT ISMYPLPAVP NKRHARVGGR KMSGSAFGKL LSRLAAEGYD 300 LSSPVDLKDH WAKHGTNRYI TIK* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
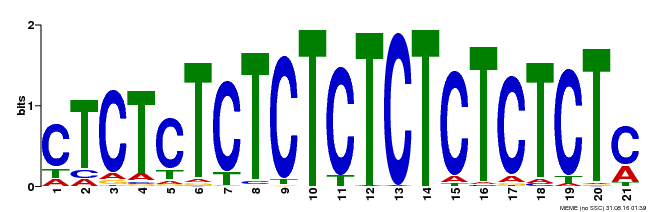
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.D01054.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010052611.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010052612.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010052613.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010052615.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010052616.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_018727567.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_018727568.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_018727569.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_018727571.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Swissprot | Q8L999 | 1e-152 | BPC6_ARATH; Protein BASIC PENTACYSTEINE6 | ||||
| TrEMBL | A0A059CE00 | 0.0 | A0A059CE00_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010052611.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6169 | 26 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-141 | basic pentacysteine 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.D01054.1.p |
| Entrez Gene | 104441262 |



