 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.D02248.1.p | ||||||||
| Common Name | EUGRSUZ_D02248, LOC104442068 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 841aa MW: 89950.2 Da PI: 6.2435 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.1 | 4.7e-21 | 127 | 182 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Eucgr.D02248.1.p 127 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 182
688999***********************************************999 PP
| |||||||
| 2 | START | 205.8 | 1.7e-64 | 340 | 569 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kae 80
ela++a++elvk+a+ +ep+W +s e++n+de++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++ + +
Eucgr.D02248.1.p 340 ELALAAMDELVKMAQTNEPLWIRSLeggrEMLNRDEYMRTFSPCIGmkpngFVTEASRETGMVIINSLALVETLMDSN-RWAEMFPcmiaRTS 431
5899**************************************9998999*9***************************.************** PP
EEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--......-TTSEE-EESSEEEEEEEECTCE CS
START 81 tlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.....sssvvRaellpSgiliepksngh 161
t +vi+sg galqlm+aelq+lsplvp R+++f+R+++q+ +g+w++vdvSvd ++ p s ++++lpSg+++++++ng+
Eucgr.D02248.1.p 432 TADVICSGmggtrnGALQLMQAELQVLSPLVPvREVTFLRFCKQHAEGVWAVVDVSVDGIRETPGggggaASFPSCCRRLPSGCVVQDMPNGY 524
************************************************************9999888874444479***************** PP
EEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 162 skvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
skvtwveh++++++++h+l+r+l++sg+ +g ++wvatlqrqc++
Eucgr.D02248.1.p 525 SKVTWVEHAEYDENHIHQLYRPLISSGMGFGSQRWVATLQRQCQC 569
*******************************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-22 | 113 | 184 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.34E-20 | 113 | 184 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 124 | 184 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 125 | 188 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.38E-18 | 126 | 184 | No hit | No description |
| Pfam | PF00046 | 1.4E-18 | 127 | 182 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 159 | 182 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 41.413 | 331 | 572 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.02E-31 | 334 | 569 | No hit | No description |
| CDD | cd08875 | 4.04E-124 | 335 | 568 | No hit | No description |
| SMART | SM00234 | 6.2E-52 | 340 | 569 | IPR002913 | START domain |
| Pfam | PF01852 | 2.2E-56 | 340 | 569 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.75E-24 | 597 | 833 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 841 aa Download sequence Send to blast |
MSFGGFLDTN SGGGGGGGGG ARVVAADIPY AGKNDMAPSG AIGQPRLLSP SLSKSMFNNS 60 PGLSLALQTG IENQQMGESY EQMSVSNHHL RRSREEEHES RSGSDNLDGG GASGDDQDAS 120 ADNPPRKKRY HRHTPQQIQE LEALFKECPH PDEKQRLELS KRLCLETRQV KFWFQNRRTQ 180 MKTQLERHEN SLLRQENDKL RAENMSLRDA MREPICSNCG GPAVIGEISL EEQHLRIENA 240 RLKDELDRVC ALAGKFLGRP VPSLAASIGP LMPNSSLELG VGTNNGFVGS LGPVAATTLP 300 LGPDFGGGIS GNSAMAVVNP SRPPGAGGGM VSIERSILLE LALAAMDELV KMAQTNEPLW 360 IRSLEGGREM LNRDEYMRTF SPCIGMKPNG FVTEASRETG MVIINSLALV ETLMDSNRWA 420 EMFPCMIART STADVICSGM GGTRNGALQL MQAELQVLSP LVPVREVTFL RFCKQHAEGV 480 WAVVDVSVDG IRETPGGGGG AASFPSCCRR LPSGCVVQDM PNGYSKVTWV EHAEYDENHI 540 HQLYRPLISS GMGFGSQRWV ATLQRQCQCL AILMSSTVSS RDHTAITPSG RRSMLKLAQR 600 MTANFCAGVC ASTVHKWNKL SGGNVDEDVR VMTRKSVDDP GEPPGIVLSA ATSVWLPVSP 660 QRLFDFLRDE QLRSEWDILS NGGPMQEMAH IAKGQDHGNC VSLLRASAMN ANQSSMLILQ 720 ETCIDAAGSL VVYAPVDIPA MHVVMNGGDS AYVALLPSGF AIVPDGPDGP GSHGGGAAPN 780 GPAASNGGPP RVGGSLLTVA FQILVNSLPT AKLTVESVET VNNLISCTIQ KIKAALQCES 840 * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
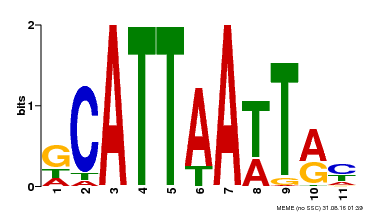
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.D02248.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010053664.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A059CIN0 | 0.0 | A0A059CIN0_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010053664.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.D02248.1.p |
| Entrez Gene | 104442068 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



