 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.D02499.1.p | ||||||||
| Common Name | EUGRSUZ_D02499, LOC104442276 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 516aa MW: 53561.5 Da PI: 8.1608 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.3 | 8.6e-16 | 328 | 373 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
Eucgr.D02499.1.p 328 HNQSERKRRDKINQRMKTLQKLVPNS-----SKTDKASVLDEVIEYLKQLQ 373
9************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.1E-18 | 321 | 389 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.784 | 323 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.17E-17 | 328 | 377 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.2E-17 | 328 | 382 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.5E-13 | 328 | 373 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.6E-18 | 329 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 516 aa Download sequence Send to blast |
MSQCVPDWDF DDSGGGGNHP TPCRRSLRSH SFNPSTPDVP MSNYEVAELT WENGQLAMHS 60 LGLPRAPAKP LPSSTAPSKY AWVDKPRAAA NAGAANAGTG TLECIVNQAT ALPRRKLPFE 120 AAAGDLVPWF DPSRGAAATA TAGTMTMDAL VPCAGAGADD RTTAHFVDSA AGSGGGVGTC 180 VVGCSTRVGS CNGAAAPGPA QEKDALAARK RARVARTAAA ATVAATAAEW SGMDQSASGS 240 ATFGRDSQQM TLDTGEMEIG AGMTSTSLGS PENTSSGKGC TRATAAADDH DSVCHSRPPR 300 EGGDDEDNKK RSGKSSVSTK RSRAAAIHNQ SERKRRDKIN QRMKTLQKLV PNSSKTDKAS 360 VLDEVIEYLK QLQAQVQMVN RMNMQPMMMP MAMQQQLQMS MMGPMGMGLG MVGMGMPGMG 420 MMDMNALGRS NLAGISPVLH PAAAAATAAA FMPMGSWGDG GDRAATATTA PMVMPDPMSP 480 FLASQSQPMT LDAYSRIAAM YAQQMHQPPP SGSKN* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 331 | 336 | ERKRRD |
| 2 | 332 | 337 | RKRRDK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
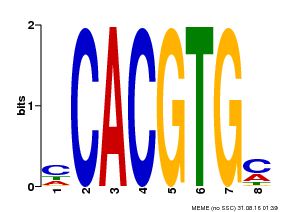
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.D02499.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010053944.1 | 0.0 | PREDICTED: transcription factor UNE10 | ||||
| TrEMBL | A0A059CK61 | 0.0 | A0A059CK61_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010053944.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3919 | 24 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 8e-63 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.D02499.1.p |
| Entrez Gene | 104442276 |



