 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.D02573.1.p | ||||||||
| Common Name | EUGRSUZ_D02573, LOC104443283 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 668aa MW: 75365.6 Da PI: 5.8064 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 156 | 2.8e-48 | 61 | 198 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspessl 94
g+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve+DGttyr++ p + +++ +s s+
Eucgr.D02573.1.p 61 GKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWTVEADGTTYRQSPPPS----QLATFSARSMDSPA 149
689******************************************************************776555....66677777777777 PP
DUF822 95 q.sslkssalaspvesy.saspksssfpspssldsislasa.......a 134
+ slks+ +++ ++s+ sa+++ ++++sp+slds+ +a++
Eucgr.D02573.1.p 150 SaASLKSYGVKTTLDSQpSAVMRMDESLSPASLDSVVIAEKdpksvkyG 198
7789*******999988467899**************998754444331 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 3.4E-49 | 61 | 204 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 3.2E-171 | 231 | 657 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 8.31E-156 | 235 | 660 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 9.7E-86 | 252 | 623 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 267 | 281 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 288 | 306 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 310 | 331 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 403 | 425 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 476 | 495 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 510 | 526 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 527 | 538 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 545 | 568 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-57 | 581 | 603 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 668 aa Download sequence Send to blast |
MNSNAGGFSD DLINNPQTDH PPDVLLPHPH LPQPPPPQQQ QQRRPRGFAA TAAAAATAVP 60 GKGKREREKE KERTKLRERH RRAITSRMLA GLRQYGNFPL PARADMNDVL AALAREAGWT 120 VEADGTTYRQ SPPPSQLATF SARSMDSPAS AASLKSYGVK TTLDSQPSAV MRMDESLSPA 180 SLDSVVIAEK DPKSVKYGNA SPINSVIECL DAEQLIHDVS SVDHQNVFTD TEFVPVYVKL 240 DGTINNYCQL VDQEVVRQEL NHIKSLNADG VVVDCWWGIV EGWNPQRYVW SGYRELFHII 300 REFKLKLQVV LAFHEYGGND SGDFSVSLPQ WVLEIGKENE DIFFADREGR RNTECLSWGI 360 DKERVLKGRT AIEVYFDLMR SFRTEFDDFF CEGLITAVEI GLGPNGELKY PSFPERMGWR 420 YPGVGEFQCY DKYLQQNLRK AAKMRGHSFW ARGPDNAGQY NSRPHETGFF CERGDYDSYY 480 GRFFLHWYAQ TLMDHADSVL SLANLAFEEA NIIVKVPAVY WWHKTASHAA ELTAGYYNPT 540 NQDGYSPVFE VLKRHSVTLK FVCLGLHSQE IDESSADPEG LNWQVLNSAW DRGLAVAGEN 600 APSCYDRDGF MRIVEMAKPR NDPDRHHFSF FVYHQPSSLV QSTMGFSELD FVIKCMRGGI 660 LEDLLPC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 233 | 620 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
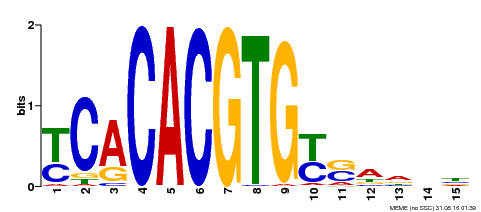
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.D02573.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010054915.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A059CJR3 | 0.0 | A0A059CJR3_EUCGR; Beta-amylase | ||||
| STRING | XP_010054915.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.D02573.1.p |
| Entrez Gene | 104443283 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



