 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.E00078.1.p | ||||||||
| Common Name | EUGRSUZ_E00078, LOC104443387 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 407aa MW: 43481.8 Da PI: 6.533 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 72 | 8.7e-23 | 261 | 323 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkrerrkq+NRe+ArrsR+RK+ae+eeL++kv++L+aeNkaLk+e+++l+++++kl+ e+
Eucgr.E00078.1.p 261 ERELKRERRKQSNRESARRSRLRKQAETEELAKKVDSLSAENKALKSEISQLTENSDKLRLEN 323
89*********************************************************9887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 3.9E-34 | 1 | 96 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.4E-12 | 115 | 235 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.3E-17 | 255 | 320 | No hit | No description |
| SMART | SM00338 | 9.1E-22 | 261 | 325 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.4E-21 | 261 | 323 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.449 | 263 | 326 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.19E-11 | 264 | 319 | No hit | No description |
| CDD | cd14702 | 4.16E-19 | 266 | 317 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 268 | 283 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 407 aa Download sequence Send to blast |
MGNNEETKSS KSDKSSSPVP PQDQTGVHVY HPDWAAMHAY YGPRVALPPY YNSAVSSGHG 60 PHPYMWGPPQ PMMPPYGPPY AAIYSHGGVY GHPAIPLTPT PLAAETPKKS SANSDNGLVK 120 KLKSFEGLAM SIGSGGDADS ADDGTDKRSS QSADSGDSSD EDQSGADKAR RKRSREGTSS 180 NGDGKSEVQG KVAGEVDAAS ENVSGGAIER PRATGKLAAP VNSPSMSSSL DLKNSCMDAN 240 ANPVSILQPG VVPPEAWLQN ERELKRERRK QSNRESARRS RLRKQAETEE LAKKVDSLSA 300 ENKALKSEIS QLTENSDKLR LENATLMERL ENAQGVEKAV ESLGKFNDNG LLSDKTENLL 360 SRVNNSGAVD RRSEDEGEIY ERKSNSGAKL HQLLDSKPRT DAVAAG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 277 | 283 | RRSRLRK |
| 2 | 277 | 284 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box-like motif (5'-ACGTGGC-3') of the chalcone synthase (CHS) gene promoter. G-box and G-box-like motifs are defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
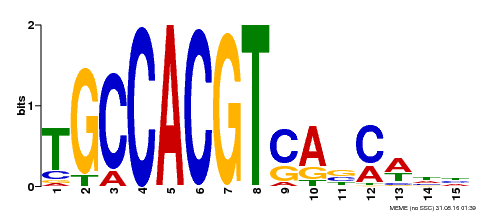
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.E00078.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010055062.1 | 0.0 | PREDICTED: common plant regulatory factor 1 isoform X3 | ||||
| Refseq | XP_018730273.1 | 0.0 | PREDICTED: common plant regulatory factor 1 isoform X1 | ||||
| Swissprot | Q99089 | 1e-132 | CPRF1_PETCR; Common plant regulatory factor 1 | ||||
| TrEMBL | A0A059C013 | 0.0 | A0A059C013_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010055062.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3571 | 26 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.1 | 2e-84 | G-box binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.E00078.1.p |
| Entrez Gene | 104443387 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



