 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.E02813.1.p | ||||||||
| Common Name | EUGRSUZ_E02813, LOC104445047 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 423aa MW: 47554.7 Da PI: 4.8541 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 108.1 | 6.7e-34 | 14 | 105 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkell 94
Fl k+ye+++ ++++ ++sws+ ++sfvv+++ efa+++LpkyFkh+nf+SF+RQLn+YgF+k++ e+ weF+++ F +g+ +ll
Eucgr.E02813.1.p 14 FLVKTYEMVDAASTDPVVSWSQGSKSFVVWNPPEFARDLLPKYFKHNNFSSFIRQLNTYGFRKIDPER---------WEFANDDFVRGQPQLL 97
9****************************************************************999.........**************** PP
XXXXXXXX CS
HSF_DNA-bind 95 ekikrkks 102
++i+r+k
Eucgr.E02813.1.p 98 KNIHRRKP 105
*****985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46785 | 8.3E-35 | 9 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 6.1E-61 | 10 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene3D | G3DSA:1.10.10.10 | 3.2E-37 | 10 | 103 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 2.8E-18 | 14 | 37 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.3E-30 | 14 | 103 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.8E-18 | 52 | 64 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 53 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.8E-18 | 65 | 77 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 423 aa Download sequence Send to blast |
MDESPSGSNS LPPFLVKTYE MVDAASTDPV VSWSQGSKSF VVWNPPEFAR DLLPKYFKHN 60 NFSSFIRQLN TYGFRKIDPE RWEFANDDFV RGQPQLLKNI HRRKPVHSHS LQNLQNQGSS 120 SPLTDAERQG LKEEVERLKG DKESLQLELQ VHEEERKGFE VQMQNLKARL LKMEERQKNM 180 VSNLGNVLQK PGLALNMLIE SEFNDKKRRL PRIGYYHDEG HGEDNHAGTP QTLTRENIDG 240 ASSLPSILDL YEQLESSLTI WEYIAHDCEN IGQRNSTLDF DETTSCADSP AISCIQFNID 300 ARPRSPTIDM NSEPTAATVF EPAVVTTSEP ALTVASEPVS SKEQAVATAP NVPAGVNDVF 360 WEQFLTENPG SSDVQEVQSE RKDTDGRRNE SKPADHSRFW WNAKTVNNLT GQLGHLTPAE 420 ST* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 3e-24 | 6 | 103 | 12 | 120 | Heat shock factor protein 1 |
| 5d5u_B | 3e-24 | 6 | 103 | 21 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 3e-24 | 6 | 103 | 21 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 3e-24 | 6 | 103 | 21 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00443 | DAP | Transfer from AT4G18880 | Download |
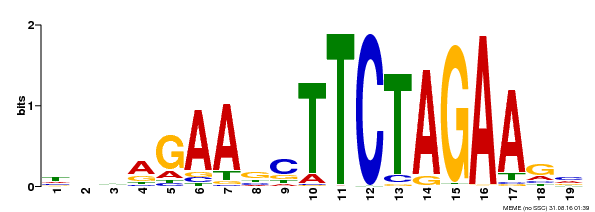
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.E02813.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010057144.1 | 0.0 | PREDICTED: heat stress transcription factor A-4c | ||||
| Swissprot | O49403 | 1e-127 | HFA4A_ARATH; Heat stress transcription factor A-4a | ||||
| TrEMBL | A0A059C7E8 | 0.0 | A0A059C7E8_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010057144.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2853 | 28 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18880.1 | 1e-121 | heat shock transcription factor A4A | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.E02813.1.p |
| Entrez Gene | 104445047 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



