 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.F00106.2.p | ||||||||
| Common Name | EUGRSUZ_F00106, LOC104447808 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 385aa MW: 43704.3 Da PI: 6.7246 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 28.6 | 2.5e-09 | 311 | 346 | 20 | 55 |
HHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 20 eknrypsaeereeLAkklgLterqVkvWFqNrRake 55
+k +yps++e+ LA+++gL+++q+ +WF N+R ++
Eucgr.F00106.2.p 311 YKWPYPSETEKVALAESTGLDQKQINNWFINQRKRH 346
4679*****************************885 PP
| |||||||
| 2 | ELK | 37.8 | 4e-13 | 266 | 287 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELK++LlrKYsgyL+sLkqE+s
Eucgr.F00106.2.p 266 ELKNHLLRKYSGYLSSLKQELS 287
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 8.7E-21 | 121 | 165 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 1.1E-22 | 122 | 164 | IPR005540 | KNOX1 |
| SMART | SM01256 | 3.5E-26 | 172 | 223 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 7.3E-23 | 177 | 221 | IPR005541 | KNOX2 |
| Pfam | PF03789 | 2.7E-10 | 266 | 287 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.296 | 266 | 286 | IPR005539 | ELK domain |
| SMART | SM01188 | 3.5E-7 | 266 | 287 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 13.005 | 286 | 349 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-20 | 287 | 362 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 8.8E-13 | 288 | 353 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-27 | 291 | 352 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 2.31E-12 | 298 | 350 | No hit | No description |
| Pfam | PF05920 | 1.5E-16 | 306 | 345 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 324 | 347 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 385 aa Download sequence Send to blast |
MEEYGQMNEN SSTGSRGNNS FLYASPVLGP SSSGNSNYGR GNSSGGHFYS QSSDHCFQSE 60 APPHPVVKTE ATTSHHGHAQ KFHHYSLVRD HHDPSASHHH HHQHHQHQQL QTASESSREV 120 DAMKAKIIAH PQYSNLLEAY MDCQKVGAPP EVVAKLSVAR QEFESRQRSS VASADGSKDP 180 ELDQFMEAYY DMLVKYRDEL TRPLQEAMDF MRRIETQLNL LSNNGPVRVF NSDEKCEGVG 240 SSEEDQDNSG GETELPEIDP RAEDRELKNH LLRKYSGYLS SLKQELSKKK KKGKLPKEAR 300 QKLLSWWELH YKWPYPSETE KVALAESTGL DQKQINNWFI NQRKRHWKPS EDMQFMVMDG 360 LHPQGAALYM DGHYIGDGPY RLGP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably binds to the DNA sequence 5'-TGAC-3'. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
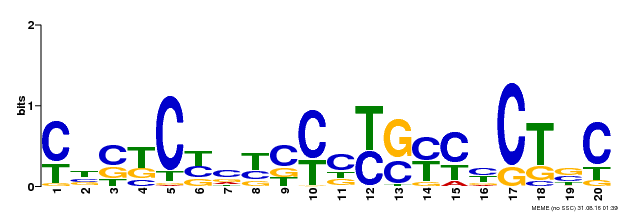
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.F00106.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010059832.1 | 0.0 | PREDICTED: homeobox protein knotted-1-like 2 | ||||
| Refseq | XP_010059833.1 | 0.0 | PREDICTED: homeobox protein knotted-1-like 2 | ||||
| Swissprot | O04135 | 0.0 | KNAP2_MALDO; Homeobox protein knotted-1-like 2 | ||||
| TrEMBL | A0A059BKP2 | 0.0 | A0A059BKP2_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010059832.1 | 0.0 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-142 | KNOTTED-like from Arabidopsis thaliana | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.F00106.2.p |
| Entrez Gene | 104447808 |



