 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.F00497.1.p | ||||||||
| Common Name | EUGRSUZ_F00497, LOC104448081 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 362aa MW: 39397.7 Da PI: 5.8268 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.1 | 1.4e-20 | 169 | 218 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ +l+g
Eucgr.F00497.1.p 169 LYRGVRQRH-WGKWVAEIRLPKN---RTRLWLGTFDTAEEAALAYDKAAYRLRG 218
59******9.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.66E-33 | 168 | 228 | No hit | No description |
| PROSITE profile | PS51032 | 23.103 | 169 | 226 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 6.2E-37 | 169 | 232 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.45E-23 | 169 | 227 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 3.0E-32 | 169 | 227 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-11 | 170 | 181 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.4E-14 | 170 | 218 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-11 | 192 | 208 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 362 aa Download sequence Send to blast |
MAAIMDFYNS ITTAALEPES FDTGGNELME ALSPFIKSAS PSNPSFSSPP CNYFSFSTPS 60 SSSSLSQPMS YPDPCSTASV TQLFPDGFSV QQSGPTGFDL YYPTQVVPQM GQIIQPQAHL 120 QDSSSYPSSF YQYHQQQQQQ QHHGGAAGFL GPRAVPMKQA GLAQKPAKLY RGVRQRHWGK 180 WVAEIRLPKN RTRLWLGTFD TAEEAALAYD KAAYRLRGDF ARLNFPHLKH KGSHIQGDFG 240 DYKPLHSSVD AKLQAICQDM AEKPADGKKR RSAPAGGGSS AAAASPRRPE PEPEPVKTEV 300 GVSAATSSSP ESDDASVEES SPLSELTFND FVEPQWESVG VPENFSLQKY PSEIDWAAIY 360 S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 8e-21 | 168 | 226 | 1 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
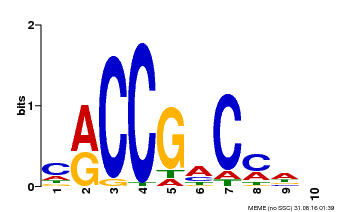
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.F00497.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010060168.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-4 | ||||
| TrEMBL | A0A059BKI8 | 0.0 | A0A059BKI8_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010060168.1 | 0.0 | (Eucalyptus grandis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2716 | 26 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78080.1 | 2e-59 | related to AP2 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.F00497.1.p |
| Entrez Gene | 104448081 |



