 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Eucgr.F02223.2.p | ||||||||
| Common Name | EUGRSUZ_F02223 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Myrtales; Myrtaceae; Myrtoideae; Eucalypteae; Eucalyptus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 519aa MW: 57509 Da PI: 7.7801 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 50.3 | 6e-16 | 181 | 240 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g ++ ++++lg ++ +e+Aa+a++ a++k++g
Eucgr.F02223.2.p 181 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqtRKgRQVYLGGYDKEEKAARAYDLAALKYWG 240
78*******************988886678446*************************98 PP
| |||||||
| 2 | AP2 | 46.1 | 1.2e-14 | 285 | 334 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
Eucgr.F02223.2.p 285 YRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 334
9***************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.32E-17 | 181 | 250 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 3.63E-22 | 181 | 248 | No hit | No description |
| Pfam | PF00847 | 3.7E-12 | 181 | 240 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.269 | 182 | 248 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.2E-29 | 182 | 254 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.5E-16 | 182 | 249 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.7E-6 | 183 | 194 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-17 | 283 | 344 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.86E-25 | 283 | 344 | No hit | No description |
| SMART | SM00380 | 7.5E-34 | 284 | 348 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.0E-18 | 284 | 342 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.177 | 284 | 342 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.1E-9 | 285 | 334 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.7E-6 | 324 | 344 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 519 aa Download sequence Send to blast |
MPLSLDSLYS YQQSADPEKQ SLDLLHEPFR HQQQHQIYYP NELTCPALYH SPLAEEEQKE 60 THIASCDENG GFSCLKSWVT RNFSSSSCHV GLDQMSSGMV CHNGGNSSEA VGPMECGDLQ 120 NLSLSMSPSS TSSCVTVPTR QISPTENECV GMEMTKKRSS NCHNKQPVHR KSIDTFGQRT 180 SQYRGVTRHR WTGRYEAHLW DNSCKKEGQT RKGRQVYLGG YDKEEKAARA YDLAALKYWG 240 PSTHINFPLE NYQKELEEMK NMSRQEYVAH LRRKSSGFSR GASMYRGVTR HHQHGRWQAR 300 IGRVAGNKDL YLGTFSTQEE AAEAYDIAAI KFRGVNAVTN FDISRYDVER IMASSTLLAG 360 ELARRTKEIA PNEEVAEYNA LTQQKIGSDG TLQLESSGNV TNGSDWKMVL YHSSPQLQAS 420 CMESSVNSSQ PSVDDTAKLG TRHLSNPSSL VTSLSSSREA SPDRAGSAFA NKPPVVSKLV 480 NTSPTSASVT SWLPSPAHMR PSAIAMSHLP VFAAWNDT* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as positive regulator of adventitious (crown) root formation by promoting its initiation. Promotes adventitious root initiation through repression of cytokinin signaling by positively regulating the two-component response regulator RR1. Regulated by the auxin response factor and transcriptional activator ARF23/ARF1. {ECO:0000269|PubMed:21481033}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
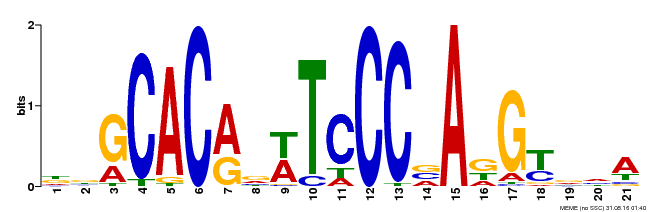
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Eucgr.F02223.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin. {ECO:0000269|PubMed:21481033}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018732061.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor ANT isoform X2 | ||||
| Swissprot | Q84Z02 | 1e-143 | CRL5_ORYSJ; AP2-like ethylene-responsive transcription factor CRL5 | ||||
| TrEMBL | A0A059BSI2 | 0.0 | A0A059BSI2_EUCGR; Uncharacterized protein | ||||
| STRING | XP_010061621.1 | 0.0 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-128 | AP2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Eucgr.F02223.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



